| |
|
|
| |
-
Time of Submission:
22:28:56 Apr 01, 2006
-
Sequence Name:
1ONF:A|PDBID|CHAIN|SEQUENCE
-
Number of residues read in:
500
-
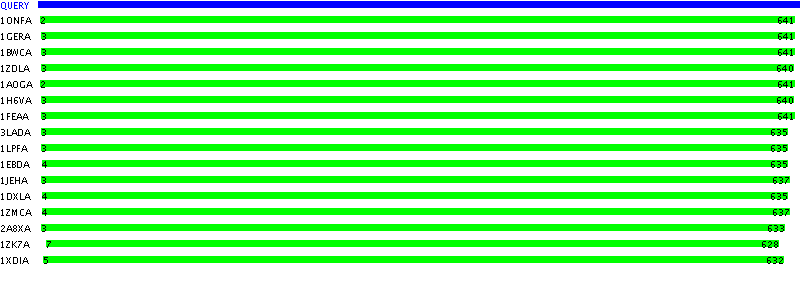
Number of useable PDB homologs found: 16
1ONFA, e-value = 0.0 CRYSTAL STRUCTURE OF PLASMODIUM FALCIPARUM GLUTATHIONE1GERA, e-value = 5.0E-97 D GLUTATHIONE REDUCTASE (E.C.1.6.4.2) COMPLEXED WITH FAD1BWCA, e-value = 5.0E-95 STRUCTURE OF HUMAN GLUTATHIONE REDUCTASE COMPLEXED WITH1ZDLA, e-value = 4.0E-71 CRYSTAL STRUCTURE OF MOUSE THIOREDOXIN REDUCTASE TYPE 21AOGA, e-value = 2.0E-66 TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE (OXIDIZED FORM)1H6VA, e-value = 8.0E-66 MAMMALIAN THIOREDOXIN REDUCTASE1FEAA, e-value = 5.0E-62 UNLIGANDED CRITHIDIA FASCICULATA TRYPANOTHIONE REDUCTASE AT3LADA, e-value = 1.0E-44 D DIHYDROLIPOAMIDE DEHYDROGENASE (E.C.1.8.1.4)1LPFA, e-value = 7.0E-43 D 2 FLAVIN-ADENINE-DINUCLEOTIDE (FAD)1EBDA, e-value = 8.0E-42 DIHYDROLIPOAMIDE DEHYDROGENASE COMPLEXED WITH THE BINDING1JEHA, e-value = 6.0E-37 CRYSTAL STRUCTURE OF YEAST E3, LIPOAMIDE DEHYDROGENASE1DXLA, e-value = 7.0E-36 DIHYDROLIPOAMIDE DEHYDROGENASE OF GLYCINE DECARBOXYLASE1ZMCA, e-value = 2.0E-32 CRYSTAL STRUCTURE OF HUMAN DIHYDROLIPOAMIDE DEHYDROGENASE2A8XA, e-value = 1.0E-30 CRYSTAL STRUCTURE OF LIPOAMIDE DEHYDROGENASE FROM1ZK7A, e-value = 9.0E-30 CRYSTAL STRUCTURE OF TN501 MERA1XDIA, e-value = 6.0E-18 CRYSTAL STRUCTURE OF LPDA (RV3303C) FROM MYCOBACTERIUM
-
Number of sequence alignments used for ab-initio predictions: 49
-
Overall confidence value:
88.8%
-
Predicted % Helix content:
33 %
(166 residues)
-
Predicted % Beta sheet content:
31 %
(157 residues)
-
Predicted % Coil content:
35 %
(177 residues)
|
| |
H = Helix
E = Beta Strand
C = Coil
Line 1 = sequence (single letter IUPAC code, 60 characters per line)
Line 2 = secondary structure (H, E or C)
Line 3 = confidence score (0-9, 0 = low, 9 = high)
A '*' character above the overall prediction indicates the homolog's structure was used at this residue.
|
| |
1 *********************************************************** 60
MVYDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDI
CCEEEEEECCCCHHHHHHHHHHHHCEEEEEEECCCCCCCCCCCCCHHHHHHHHHHHHHHH
999999999999999999999999999999999999999999999999999999999999
61 ************************************************************ 120
LENSRHYGFDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASFLSENRIL
HHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCEEEEEEEEEEECCEEEE
999999999999999999999999999999999999999999999999999999887777
121 ************************************************************ 180
IKGTKDNNNKDNGPLNEEILEGRNILIAVGNKPVFPPVKGIENTISSDEFFNIKESKKIG
EEECCCCCCCCEEEEEEEEEEEEEEEEEEEEEEECCCCCCCCCEEECHHHHCCCCCEEEE
997677888887787778889999999999999999999999999999999999999999
181 ************************************************************ 240
IVGSGYIAVELINVIKRLGIDSYIFARGNRILRKFDESVINVLENDMKKNNINIVTFADV
EECCCHHHHHHHHHHHHCCEEEEEEEEEECCCCCCCHHHHHHHHHHHHHHCEEEEEEEEE
999999999999999999999999999999999999999999999999999999999999
241 ************************************************************ 300
VEIKKVSDKNLSIHLSDGRIYEHFDHVIYCVGRSPDTENLKLEKLNVETNNNYIVVDENQ
EEEEECCCCEEEEEECCCEEEEEEEEEEEECCEEEECCCCCCCCCCEEECCCEEEEECCC
999999999999999999999999999999999999999999999999999999999999
301 ********************************************* ************* 360
RTSVNNIYAVGDCCMVKKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINA
CCCCCCEEEECCEEEECHHHHHHHCCCCHHHHHHHHHHHHHHEEECCCCCCCCHHHHHHH
999999999999999997377777888799999999999998777779999999999999
361 ************************************************************ 420
GRLLADRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNL
HHHHHHHHCCCCCCCCCCCCCEEEECCCEEEEEECCCHHHHHHHHCCCEEEEEEEEEECC
999999999999999999999999999999999999999999999999999999999999
421 ************************************************************ 480
FFSVYDIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDE
CCCCCCCCCCCCCEEEEEEEEECCCCCCEEEEEEECCHHHHHHHHHHHHHHCCCHHHHHH
999999999999999999999999999999999999999999999999999999999999
481 ******************** 500
TIPIHPTAAEEFLTLQPWMK
CCCCCCCCHHHCCCCCCCCC
99999999999999998889
|
|
|
| |
|
1 MVYDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDI 60
CCEEEEEECCCHHHHHHHHHHHHCCCCEEEEECCCCCCEEEECCCCCCHHHHCHHHHHHH
965699998998999999999987997999988988877888677776455445778999
61 LENSRHYGFDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASFLSENRIL 120
HHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCEEEEEEEEEEECCEEE
986676687765554699999999999999999999999869858999999998585999
121 IKGTKDNNNKDNGPLNEEILEGRNILIAVGNKPVFPPVKGIENTISSDEFFNIKESKKIG 180
EEECCCCCCEEECCCCCEEEEECEEEEECCCEEEECCCCCHHHHCCCHHHHHHHCCCEEE
986677655455578875998788999689865558899855654657887665789799
181 IVGSGYIAVELINVIKRLGIDSYIFARGNRILRKFDESVINVLENDMKKNNINIVTFADV 240
EECCCHHHHHHHHHHHHCCCEEEEEECCCCHHHHCCHHHHHHHHHHHHHCCCEEEECCEE
996888999999999987996999976884455656999999999999789999989899
241 VEIKKVSDKNLSIHLSDGRIYEHFDHVIYCVGRSPDTENLKLEKLNVETNNNYIVVDENQ 300
EEEEECCCCEEEEEECCCEEEEEEEEEEEEECEEECHHHCCCCCCCEEECCCEEEECCCC
999996898999999799588776799998575546665466777678779978989846
301 RTSVNNIYAVGDCCMVKKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINA 360
CCCCCCEEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHH
679998599876346766544455667889999998765655455556788765899999
361 GRLLADRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNL 420
HHHHHHHHHCCCCCCCCCCCEEEEEEEEEEEEEEECCHHHHHHHCCCCCEEEEEECCCCC
999999985998766788866899996645788868999999965988669998657877
421 FFSVYDIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDE 480
CCCCCCCCCCCCCCEEEEEEEECCCCEEEEEEEECCCHHHHHHHHHHHHHCCCCHHHHHH
665555444689985899999999999999999869989999999999988999999984
481 TIPIHPTAAEEFLTLQPWMK 500
HCCCCCCCHHHHHHHCCCCC
46778884676886556679
1 MVYDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDI 60
CCCEEEEECCCCCHHHHHHHHHHCCCCEEEEEECCCCCEEEEECCCCCCHHHHHHHHHHH
636168871785147889999870784588871888642686143567012334688899
61 LENSRHYGFDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASFLSENRIL 120
HHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCEEEEEEEEEEECCCEEE
987358887645310489999998888876421156654118817887235674575478
121 IKGTKDNNNKDNGPLNEEILEGRNILIAVGNKPVFPPVKGIENTISSDEFFNIKESKKIG 180
EEECCCCCEEEECCCCCEEEEEEEEEEECCCCCCCCCCCCCCCCCCHHHHHHCCCCCCEE
871688734775068860553013677558876789998752101102455307999278
181 IVGSGYIAVELINVIKRLGIDSYIFARGNRILRKFDESVINVLENDMKKNNINIVTFADV 240
EECCCHHHHHHHHHHHHCCCCEEEEEECCCCCCCCCHHHHHHHHHHHHHCCCEEEECCCE
870460456789998864994688762887756650678888888887489968863513
241 VEIKKVSDKNLSIHLSDGRIYEHFDHVIYCVGRSPDTENLKLEKLNVETNNNYIVVDENQ 300
EEEEECCCCEEEEEECCCCCEEEEEEEEEEECCCCCCCCHHHHHHHHCCCCCEEEEECCC
688865885688887278532212356777314167211133232111288728874145
301 RTSVNNIYAVGDCCMVKKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINA 360
CCCCCCEEEECCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHH
667994488603655553123456789999989999998776533436676315888888
361 GRLLADRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNL 420
HHHHHHHHHCCCCCCCCCCCCCEEEEECCCCEECCCHHHHHHHHCCCCCEEEEEECCCCC
999999982788776788762068883442121365268887761886178874137841
421 FFSVYDIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDE 480
HHHHHHHHHCCCCCCEEEEEEECCCCEEEEEEEECCCCHHHHHHHHHHHHCCCCHHHHHC
111001233799853478888579963678887358862789999999862675466512
481 TIPIHPTAAEEFLTLQPWMK 500
CCCCCCCCHHHHHHHCCCCC
33556775467764189899
1 MVYDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDI 60
CCCEEEEECCCCCCHHHHHHHHHHCEEEEEEECCCCCEEEEEEEECCCCEEEEEEEEEEE
987899985799866788777766979999996778968999876576446777778776
61 LENSRHYGFDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASFLSENRIL 120
ECCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHCCCEEEEEEEEEEEECCEEEECCCCE
568767898766665457788877777766555445546655689998575788655668
121 IKGTKDNNNKDNGPLNEEILEGRNILIAVGNKPVFPPVKGIENTISSDEFFNIKESKKIG 180
EEEEECCCCCEECCCCCCEEEEEEEEEEECCCCCCCCCCCCCEEEEECHHHHHCCCCEEE
999776898654677889899999999976776787678754676555456645687699
181 IVGSGYIAVELINVIKRLGIDSYIFARGNRILRKFDESVINVLENDMKKNNINIVTFADV 240
EECCCCEEEECHHHHHHCCEEEEEEEECHHHHCCCCCHHHHHHHHHHHCCEEEEEEEEEE
976875565555577748847898887535447887557899888654468988667899
241 VEIKKVSDKNLSIHLSDGRIYEHFDHVIYCVGRSPDTENLKLEKLNVETNNNYIVVDENQ 300
EEEEECCCCCEEECCEEEECCCCEEEEEEEECCCCCCCCEEEEEEEEEECCCEEEEEEEC
999876899789966876688988999999858888775445678888758899999865
301 RTSVNNIYAVGDCCMVKKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINA 360
CCEEEEEEEECCCEEEEEEEEEEEEEEEEEEEEEEEEECCCCCEEECCCCCEEEHHHHHH
885678999867545778899999999999999999988898777656774455567655
361 GRLLADRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNL 420
HHHHCEEEEECCCCEEEEEEEEEEEECCCCCCCCCHHHHHHHHHCCCCEEEEEEEEEEEE
466545565368865665678889875777656655777777766997589999998665
421 FFSVYDIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDE 480
EECCCCCCCCCCCCEEEEEEEEECCCEEEEEEEEECCCHHHHHHHHHHHHHCCEEEEEEE
445776544577766799995866885889999865877789999998765674566655
481 TIPIHPTAAEEFLTLQPWMK 500
EEECCCCCCHHHHHHCCCCC
56658887557886468889
1 MVYDLIVIGGGSGGMAAARRAARHNAKVALVEKSRLGGTCVNVGCVPKKIMFNAASVHDI 60
CEEEEEEECCCCHHHHHHHHHHHHCCEEEEEEECCCCEEEEEECCCCCHHHHHHHHHHHH
969999975896799999999984868999985899869998667777688889999999
61 LENSRHYGFDTKFSFNLPLLVERRDKYIQRLNNIYRQNLSKDKVDLYEGTASFLSENRIL 120
HHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCEEEEEEEEEEEECCCEEE
998765788888876489999999999999999999999866699999999997797689
121 IKGTKDNNNKDNGPLNEEILEGRNILIAVGNKPVFPPVKGIENTISSDEFFNIKESKKIG 180
EEECCCCCEEEECCCCEEEEEEEEEEEECCCCEEECCCCCHHHCCCHHHHHHHCCCEEEE
999689875787689958999899999867767756777645446747889865896999
181 IVGSGYIAVELINVIKRLGIDSYIFARGNRILRKFDESVINVLENDMKKNNINIVTFADV 240
EEECCHHHHHHHHHHHHCCEEEEEEEECCCCCCCCCHHHHHHHHHHHHHCCEEEEECCEE
964877899999999885879999996776455687899999999999865999977778
241 VEIKKVSDKNLSIHLSDGRIYEHFDHVIYCVGRSPDTENLKLEKLNVETNNNYIVVDENQ 300
EEEEECCCCEEEEEECCCCCEEEEEEEEEEECCCCCCCCCCCCCEEEEECCEEEEEECCC
999967997799997578766889999999655567766565544668768699995678
301 RTSVNNIYAVGDCCMVKKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINA 360
CCCCCEEEEEEEECCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHH
887758998876567776378999999999999999999998646556888755999999
361 GRLLADRLFLKKTRKTNYKLIPTVIFSHPPIGTIGLSEEAAIQIYGKENVKIYESKFTNL 420
HHHHHHHHHCCCCCCCCCCEEEEEEEEECCEEEECCCHHHHHHHCCCCEEEEEEEECCCC
999999998477877766658999998566567656589999975897899999957775
421 FFSVYDIEPELKEKTYLKLVCVGKDELIKGLHIIGLNADEIVQGFAVALKMNATKKDFDE 480
CCCCCCCCCCCCCCEEEEEEEECCCCEEEEEEEECCCHHHHHHHHHHHHHHCCCHHHHHH
567777766788868999999879988999999769959999999999996553799986
481 TIPIHPTAAEEFLTLQPWMK 500
HCCCCCCCHHHHHHHCCCCC
45888986799988768889
QUERY
MVYDLIVIGGGSGGMAAARRAAR-HN---AKVALVE---------K---------S---- 34
1ONFA
~VYDLIVIGGGSGGMAAARRAAR-HN---AKVALVE---------K---------S----
~CEEEEEECCCCHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
1GERA
~~YDYIAIGGGSGGIASINRAAM-YG---QKCALIE---------A---------K----
~~EEEEEECCHHHHHHHHHHHHH~CC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
1BWCA
~~YDYLVIGGGSGGLASARRAAE-LG---ARAAVVE---------S---------H----
~~EEEEEECCCHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~E~~~~
1ZDLA
~~FDLLVIGGGSGGLACAKEAAQ-LG---KKVAVADYVEPSPRGTK---------W----
~~EEEEEECCCHHHHHHHHHHHH~HC~~~EEEEEEEECCCCCCCCC~~~~~~~~~C~~~~
1AOGA
~IFDLVVIGAGSGGLEAAWNAATLYK---KRVAVID---------VQMVHGPPFFS----
~EEEEEEECCCHHHHHHHHHHHHCCC~~~EEEEEEE~~~~~~~~~CCCCCCCCCCC~~~~
1H6VA
~~FDLIIIGGGSGGLAAAKEAAK-FD---KKVMVLDFVTPTPLGTN---------W----
~~EEEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEEECCCCCCCCC~~~~~~~~~E~~~~
1FEAA
~~YDLVVIGAGSGGLEAGWNAASLHK---KRVAVID---------L---------Q----
~~EEEEEECCCCHHHHHHHHHHHCCC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
3LADA
~~FDVIVIGAGPGGYVAAIKSAQ-LG---LKTALIE---------K---------Y----
~~EEEEEECCCHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~E~~~~~~~~~E~~~~
1LPFA
~~FDVVVIGAGPGGYVAAIRAAQ-LG---LKTACIE---------K---------YIGKE
~~EEEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~E~~~~~~~~~CCCCC
1EBDA
~~~ETLVVGAGPGGYVAAIRAAQ-LG---QKVTIVE---------K---------G----
~~~EEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
1JEHA
~~HDVVIIGGGPAGYVAAIKAAQ-LG---FNTACVE---------KR--------G----
~~EEEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~CC~~~~~~~~C~~~~
1DXLA
~~~DVVIIGGGPGGYVAAIKAAQ-LG---FKTTCIE---------KR--------G----
~~~EEEEECCCHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~CC~~~~~~~~C~~~~
1ZMCA
~~~DVTVIGSGPGGYVAAIKAAQ-LG---FKTVCIE---------K---------N----
~~~EEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
2A8XA
~~YDVVVLGAGPGGYVAAIRAAQ-LG---LSTAIVE---------P---------K----
~~EEEEEECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
1ZK7A
~~~~~~VIGSGGAAMAAALKAVE-QG---AQVTLIE---------R---------G----
~~~~~~EECCHHHHHHHHHHHHH~HC~~~EEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
1XDIA
~~~~IVILGGGPAGYEAALVAAT-SHPETTQVTVID---------C---------D----
~~~~EEEECCCHHHHCHHHHHHH~HCCCEEEEEEEE~~~~~~~~~C~~~~~~~~~C~~~~
34 ---R---------LGGTCVNVGCVPKK-IMFNAA-SV-HDILE-NSRHYGF--D---T-- 71
1ONFA
---R---------LGGTCVNVGCVPKK-IMFNAA-SV-HDILE-NSRHYGF--D---T--
~~~C~~~~~~~~~CCCCCCCCCCHHHH~HHHHHH~HH~HHHHH~CHHHCCC~~C~~~C~~
1GERA
---E---------LGGTCVNVGCVPKK-VMWHAA-QI-REAIHMYGPDYGF--D---T--
~~~C~~~~~~~~~CCCCCCCCCHHHHH~HHHHHH~HH~HHHHHHHHHHHCC~~C~~~C~~
1BWCA
---K---------LGGTCVNVGCVPKK-VMWNTA-VH-SEFMH-DHADYGF--P---SC-
~~~E~~~~~~~~~ECCCCCCCCHHHHH~HHHHHH~HH~HHHHH~HHCCCCC~~C~~~CC~
1ZDLA
---G---------LGGTCVNVGCIPKK-LMHQAA-LL-GGMIR-DAHHYGW--E---VAQ
~~~C~~~~~~~~~CCCEEECCCCHHHH~HHHHHH~HH~HHHHH~HHHCCCC~~C~~~CCC
1AOGA
---A---------LGGTCVNVGCVPKK-LMVTGA-QY-MEHLR-ESAGFGW--EFDRT--
~~~C~~~~~~~~~CCCCCCCCCHHHHH~HHHHHH~HH~HHHHH~HHHHCCC~~CCCCC~~
1H6VA
---G---------LGGTCVNVGCIPKK-LMHQAA-LL-GQALK-DSRNYGWKLE---D--
~~~E~~~~~~~~~ECCCCCCCCHHHHH~HHHHHH~HH~HHHHH~HHHHHCCCCC~~~C~~
1FEAA
---KHHGPPHYAALGGTCVNVGCVPKK-LMVTGA-NY-MDTIR-ESAGFGW--ELDRE--
~~~CCCCCCCCCCCCCCCCCCCHHHHH~HHHHHH~HH~HHHHH~HHHCCCC~~CCCCC~~
3LADA
---KGKEGKTA--LGGTCLNVGCIPSK-ALLDSSYKF-HEAHE-SFKLHGI--S---TG-
~~~CCCCCCEE~~ECCCCCCCCCHHHH~HHHHHHHHH~HHHHC~CCCCCCC~~C~~~CC~
1LPFA
GKVA---------LGGTCLNVGCIPSK-ALLDSSYKY-HEAKE-AFKVHGI--E---A--
CCEE~~~~~~~~~ECCHHHHCCHHHHH~HHHHHHHHH~HHHHH~CCCCCCC~~C~~~C~~
1EBDA
---N---------LGGVCLNVGCIPSK-ALISAS-HR-YEQAK-HSEEMGI--K---A--
~~~C~~~~~~~~~CCCCCCCCCCHHHH~HHHHHH~HH~HHHHH~CCCCCCC~~C~~~C~~
1JEHA
---K---------LGGTCLNVGCIPSK-ALLNNS-HLFHQMHT-EAQKRGI--D---V--
~~~C~~~~~~~~~CCCCCCCCCHHHHH~HHHHHH~HHHHHHHH~HHHCCCC~~C~~~C~~
1DXLA
---A---------LGGTCLNVGCIPSK-ALLHSS-HMYHEAKH-SFANHGV--KV--S--
~~~C~~~~~~~~~CCCEEECCCCCHHH~HHHHHH~HHHHHHHH~HCCCCCC~~CC~~C~~
1ZMCA
---ET--------LGGTCLNVGCIPSK-ALLNNS-HY-YHMAH-GTDFASR--G---I--
~~~CC~~~~~~~~CCCHHHHCCHHHHH~HHHHHH~HH~HHHHH~CHHHHHH~~C~~~C~~
2A8XA
---Y---------WGGVCLNVGCIPSKALLRNAE-LV-HIFTK-DAKAFGI--S---G--
~~~C~~~~~~~~~CCCCCCCCCCHHHHHHHHHHH~HH~HHHHH~CCCCCCC~~C~~~C~~
1ZK7A
---T---------IGGTCVNVGCVPSK-IMIRAA-HI-AHL----RRESPF--D---G--
~~~C~~~~~~~~~CCCHHHHCCHHHHH~HHHHHH~HH~HHH~~~~HHCCCC~~C~~~C~~
1XDIA
---G---------IGGAAVLDDCVPSK-TFIAST-GL-RTELR-RAPHLGF--H------
~~~C~~~~~~~~~CCCCCCCCHHHHHH~HHHHHH~HH~HHHCC~CHHHHCC~~C~~~~~~
72 K--FS---FNLPL-----LVERRDKYIQ-RLNNIYRQNL--S-KDKVDLYEGTASFLSEN 117
1ONFA
K--FS---FNLPL-----LVERRDKYIQ-RLNNIYRQNL--S-KDKVDLYEGTASFL---
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~CCCEEEEEEECEEE~~~
1GERA
T--INK--FNWET-----LIASRTAYID-RIHTSYENVL--G-KNNVDVIKGFARFVDAK
C~~CCC~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEECCCE
1BWCA
E--GK---FNWRV-----IKEKRDAYVS-RLNAIYQNNL--T-KSHIEIIRGHAAFTSDP
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEECEEEEECCC
1ZDLA
P--VQ---HNWKT-----MAEAVQNHVK-SLNWGHRVQL--Q-DRKVKYFNIKASFVDEH
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHCHHHHH~~H~CCCEEEEEEEEEECCCC
1AOGA
T--LR---AEWKN-----LIAVKDEAVL-NINKSYDEMFR-D-TEGLEFFLGWGSLESKN
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHHH~H~CCCEEEEECEEEECCCE
1H6VA
T--VK---HDWEK-----MTESVQNHIG-SLNWGYRVAL--R-EKKVVYENAYGKFIGPH
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEECCCC
1FEAA
S--VR---PNWKA-----LIAAKNKAVS-GINDSYEGMF--ADTEGLTFHQGFGALQDNH
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~HCCCCEEEEECEEEECCCE
3LADA
E--VA---IDVPT-----MIARKDQIVR-NLTGGVASLI--K-ANGVTLFEGHGKLLAGK
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEEECCC
1LPFA
KG-VT---IDVPA-----MVARKANIVK-NLTGGIATLF--K-ANGVTSFEGHGKLLANK
CC~CC~~~CHHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEEECCC
1EBDA
EN-VT---IDFAK-----VQEWKASVVK-KLTGGVEGLL--K-GNKVEIVKGEAYFVDAN
CC~CC~~~CHHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HCCEEEEEEEEEECCCE
1JEHA
NGDIK---INVAN-----FQKAKDDAVK-QLTGGIELLF--K-KNKVTYYKGNGSFEDET
CCCCC~~~CHHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEECCCC
1DXLA
N--VE---IDLAA-----MMGQKDKAVS-NLTRGIEGLF--K-KNKVTYVKGYGKFVSPS
C~~CC~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEECCCE
1ZMCA
E--MSEVRLNLDK-----MMEQKSTAVK-ALTGGIAHLF--K-QNKVVHVNGYGKITGKN
C~~CCCCCCHHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEECCCE
2A8XA
E--VT---FDYGI-----AYDRSRKVAEGRVAGVH--FL--K-KNKITEIHGYGTFADAN
C~~CC~~~CCHHH~~~~~HHHHHHHHHHHHHHHHH~~HC~~C~CCCEEEEEEEEEECCCE
1ZK7A
G--IA---ATVPTIDRSKLLAQQQARVD-ELRHAKYEGILGG-NPAITVVHGEARFKDDQ
C~~CC~~~EEECCCCHHHHHHHHHHHHH~HHHHHHCHHHHCC~CCCEEEEEEEEEECCCE
1XDIA
----K---ISLPQ-----IHARVKTLAA-AQSADITAQL--L-SMGVQVIAGRGELIDST
~~~~C~~~CCHHH~~~~~HHHHHHHHHH~HHHHHHHHHH~~H~HHCEEEEEEEEEEECCC
118 RILIKGTKDNNNKDNGPLNEEILEGRNILIAVGNKP-V-FP---PVKG-I-E----NT-- 164
1ONFA
-----------------------EGRNILIAVGNKP-V-FP---PVKG-I-E----NT--
~~~~~~~~~~~~~~~~~~~~~~~CEEEEEEEECEEE~E~CC~~~CCCC~C~C~~~~CE~~
1GERA
TLEVNG--------------ETITADHILIATGGRP-S-HP---DIPG-V-E----YG--
EEEECC~~~~~~~~~~~~~~CEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CE~~
1BWCA
KPTIEVS---GKKYTAP---------HILIATGGMP-S-TPHESQIPG-A-S----LG--
CEEEEEC~~~CEEEEEC~~~~~~~~~EEEEECCEEE~E~CCCHHHCCC~C~C~~~~CE~~
1ZDLA
TV--------RGVDKGG-KATLLSAEHIVIATGGRP-R-YPT--QVKGAL-E----YG--
EE~~~~~~~~EEEECCC~EEEEEEEEEEEEECCEEE~E~CCC~~CCCCCC~C~~~~CE~~
1AOGA
VVNVR-----ESADPASAVKERLETEHILLASGSWP-H-MP---NIPG-I-E----HC--
EEEEE~~~~~ECCCCCEEEEEEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CE~~
1H6VA
KIMA-----TNNKG----KEKVYSAERFLIATGERP-R-YL---GIPGDK-E----YC--
EEEE~~~~~ECCCC~~~~EEEEEEEEEEEEECCEEE~E~CC~~~CCCCCC~C~~~~CE~~
1FEAA
TVLVRESADPNS-----AVLETLDTEYILLATGSWP-Q-HL---GIEG-D-D----LC--
EEEEEECCCCCE~~~~~EEEEEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CE~~
3LADA
KVEVTAADGSS---------QVLDTENVILASGSKP-V-EI---PPAP-V-D----QD--
EEEEECCCCEE~~~~~~~~~EEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CC~~
1LPFA
QVEVTGLDGKT---------QVLEAENVIIASGSRP-V-EI---PPAP-L-S----DD--
EEEEECCCCEE~~~~~~~~~EEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CC~~
1EBDA
TVRVV---------NGD-SAQTYTFKNAIIATGSRP-IELP---NFKF-S-N----RI--
EEEEE~~~~~~~~~ECC~CEEEEEEEEEEEECCEEE~ECCC~~~CCCC~C~C~~~~EE~~
1JEHA
KIRVTPV---DGLEGTVKEDHILDVKNIIVATGSEV-T------PFPG-I-EIDEEKI--
EEEEEEC~~~CCCCCCCCCCEEEEEEEEEEEECEEE~E~~~~~~CCCC~C~CCCCCEE~~
1DXLA
EISVDTIEGENT---------VVKGKHIIIATGSDV-K-SL---PGVT-I-DEK--KI--
EEEEECCCCCEE~~~~~~~~~EEEEEEEEEECCEEE~E~CC~~~CCCC~C~CCC~~EE~~
1ZMCA
QVTA------TKADGG---TQVIDTKNILIATGSEV-TPFP---GITI-D-E----DT--
EEEE~~~~~~ECCCCE~~~EEEEEEEEEEEECCEEE~ECCC~~~CCCC~C~C~~~~CE~~
2A8XA
TLLV------DLNDGG---TESVTFDNAIIATGSST-R-LV---PGTS-L-SA---NV--
EEEE~~~~~~EECCCE~~~EEEEEEEEEEEECCEEE~E~CC~~~CCCC~C~CC~~~CE~~
1ZK7A
SLTVRLNEG---------GERVVMFDRCLVATGASP-A-VP---PIPG-L-K----ESPY
EEEEEECCC~~~~~~~~~CEEEEEEEEEEEECCEEE~E~CC~~~CCCC~C~C~~~~CCCE
1XDIA
PGLARHRIKATAAD-GSTSEH--EADVVLVATGASPRI-LP---SAQP-DGE----RI--
CCCCEEEEEEECCC~CEEEEE~~EEEEEEEECCEEEEC~CC~~~CCCC~CCC~~~~EE~~
164 -I-SSD-----EFFNIKE-S---K-KIGI-VGSGYIAVE---LINVIKRLGID-SYIFAR 207
1ONFA
-I-SSD-----EFFNIKE-S---K-KIGI-VGSGYIAVE---LINVIKRLGID-SYIFAR
~E~ECH~~~~~HHHCCCC~C~~~E~EEEE~ECCCHHHHH~~~HHHHHHHCCEE~EEEEEE
1GERA
-I-DSD-----GFFALPALP---E-RVAV-VGAGYIAVE---LAGVINGLGAK-THLFVR
~E~ECH~~~~~HHCCCCCCC~~~E~EEEE~ECCCHHHHH~~~HHHHHHHHCEE~EEEEEE
1BWCA
-I-TSD-----GFFQLEE-L---PGRSVI-VGAGYIAVE---MAGILSALGSK-TSLMIR
~E~EHH~~~~~HHCCCCC~C~~~CEEEEE~ECCHHHHHH~~~HHHHHHHHCEE~EEEEEE
1ZDLA
-I-TSD-----DIFWLKE-S---PGKTLV-VGASYVALE---CAGFLTGIGLD-TTVMMR
~E~ECC~~~~~HHHCCCC~C~~~CCEEEE~EECCCHHHH~~~HHHHHHHHCEE~EEEEEC
1AOGA
-I-SSN-----EAFYLPEPP---R-RVLT-VGGGFISVEFAGIFNAYKPKDGQ-VTLCYR
~E~EHH~~~~~HHHCCCCCC~~~E~EEEE~ECCHHHHHHHHHHHHHCCCCCEE~EEEEEE
1H6VA
-I-SSD-----DLFSLPY-C---PGKTLV-VGASYVALE---CAGFLAGIGLD-VTVMVR
~E~ECH~~~~~HHHCCCC~C~~~CCEEEE~ECCHHHHHH~~~HHHHHHHHCEE~EEEEEC
1FEAA
-I-TSN-----EAFYLDE-A---P-KRALCVGGGYISIEFAGIFNAYKARGGQ-VDLAYR
~E~EHH~~~~~HHHCCCC~C~~~C~EEEEEECCHHHHHHHHHHHHHHCCCCEE~EEEEEE
3LADA
-V-IVDSTGALDFQNV---P---G-KLGV-IGAGVIGLE---LGSVWARLGAE-VTVLEA
~E~EEECHHHHCCCCC~~~C~~~E~EEEE~ECCHHHHHH~~~HHHHHHHHCEE~EEEEEE
1LPFA
-I-IVD-----STGALEF-QAVPK-KLGV-IGAGVIGLE---LGSVWARLGAE-VTVLEA
~C~EEE~~~~~HHHHHCC~CCCCE~EEEE~ECCCHHHHH~~~HHHHHHHHCEE~EEEEEE
1EBDA
-L-DST-----GALNLGEVP---K-SLVV-IGGGYIGIE---LGTAYANFGTK-VTILEG
~E~EHH~~~~~HHHHCCCCC~~~E~EEEE~ECCHHHHHH~~~HHHHHHHHCEE~EEEEEE
1JEHA
-V-SST-----GALSLKEIP---K-RLTI-IGGGIIGLE---MGSVYSRLGSK-VTVVEF
~E~CHH~~~~~HHHCCCCCC~~~E~EEEE~ECCCHHHHH~~~HHHHHHHHCEE~EEEEEC
1DXLA
-V-SST-----GALALSEIP---K-KLVV-IGAGYIGLE---MGSVWGRIGSE-VTVVEF
~E~CHH~~~~~HHHCCCCCC~~~E~EEEE~EECCHHHHH~~~HHHHHHHHCEE~EEEEEC
1ZMCA
-IVSST-----GALSLKKVP---E-KMVV-IGAGVIGVE---LGSVWQRLGADVTAVEFL
~EEEHH~~~~~HHHCCCCCC~~~E~EEEE~ECCCHHHHH~~~HHHHHHHHCEEEEEEEEE
2A8XA
-V-TYE-----EQILSRELP---K-SIII-AGAGAIG-E---FGYVLKNYGVD-VTIVEF
~E~EHH~~~~~HHCCCCCCC~~~E~EEEE~ECCCCCC~C~~~HHHHHHHHCEE~EEEEEE
1ZK7A
WT-STE-----ALASDTI-P---E-RLAV-IGSSVVALE---LAQAFARLGSK-VTVLAR
EE~HHH~~~~~HHHCCCC~C~~~E~EEEE~ECCCHHHHH~~~HHHHHHHHCEE~EEEEEC
1XDIA
-L-TWR-----QLYDLDALP---D-HLIV-VGSGVTGAE---FVDAYTELGVP-VTVVAS
~E~ECC~~~~~CCCCCCCCC~~~C~EEEE~ECCCCCHHH~~~HHHHHHCCCEE~EEEEEE
208 GNRILRKF-DESVINVLENDMKKNNINIV-----TFADVVEI-----KK-VSD-K--NL- 251
1ONFA
GNRILRKF-DESVINVLENDMKKNNINIV-----TFADVVEI-----KK-VSD-K--NL-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~C~~EE~
1GERA
KHAPLRSF-DPMISETLVEVMNAEGPQLH-----TNAIPKAV-----VK-NTD-G--SL-
ECCCCCCC~CHHHHHHHHHHHHHCCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~C~~EE~
1BWCA
HDKVLRSF-DSMISTNCTEELENAGVEVL-----KFSQVKEV-----KKTLSG-L--EV-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EECCCC~E~~EE~
1ZDLA
-SIPLRGF-DQQMSSLVTEHMESHGTQFL-----KGCVPSHI-----KK-LPT-N--QL-
~CCCCCCC~HHHHHHHHHHHHHHCCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~C~~EE~
1AOGA
GEMILRGF-DHTLREELTKQLTANGIQIL-----TKENPAKV-----EL-NAD-G--SK-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECCCEEEE~~~~~EE~CCC~C~~EE~
1H6VA
-SILLRGF-DQDMANKIGEHMEEHGIKFIRQFVPTKIEQIEAGTPGRLK-VTA-K--ST-
~CCCCCCC~CHHHHHHHHHHHHHHCEEEEECEEEEEEEEEECCCCCEEE~EEE~E~~CC~
1FEAA
GDMILRGF-DSELRKQLTEQLRANGINVR-----THENPAKV-----TK-NAD-G--TR-
ECCCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECCCEEEE~~~~~EE~CCC~C~~EE~
3LADA
MDKFLPAV-DEQVAKEAQKILTKQGLKIL-----LGARVTG------TE-VKN-K--QV-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEE~~~~~~EE~ECC~C~~EE~
1LPFA
LDKFLPAA-DEQIAKEALKVLTKQGLNIR-----LGARVT-A-----SE-VKK-K--QV-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEE~E~~~~~EE~ECC~C~~EE~
1EBDA
AGEILSGF-EKQMAAIIKKRLKKKGVEVV-----TNALAKGA-----EE-RED-G--VT-
ECCCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~C~~EE~
1JEHA
QPQIGASM-DGEVAKATQKFLKKQGLDFK-----LSTKVISA-----KR-NDD-K--NVV
CCCCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~ECC~C~~CEE
1DXLA
ASEIVPTM-DAEIRKQFQRSLEKQGMKFK-----LKTKVVGV-----DT-SGDGV--KL-
CCCCCCCC~CHHHHHHHHHHHHCCCEEEE~~~~~ECEEEEEE~~~~~EE~CCCEE~~EE~
1ZMCA
GHVGGVGI-DMEISKNFQRILQKQGFKFK-----LNTKVTGA-----TK-KSD-GKIDV-
ECCCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~CEEEE~
2A8XA
LPRALPNE-DADVSKEIEKQFKKLGVTIL-----TATKVESI-----AD-GGS-Q--VT-
ECCCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~CCC~E~~EE~
1ZK7A
NTLFFRE--DPAIGEAVTAAFRAEGIEVL-----EHTQASQV-----AH-M-D-G--EF-
CCCCCCC~~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~C~C~C~~EE~
1XDIA
QDHVLPYE-DADAALVLEESFAERGVRLF-----KNARAASV-----TR-TG--A--GV-
EECCCCCC~CHHHHHHHHHHHHHHCEEEE~~~~~ECEEEEEE~~~~~EE~CC~~C~~EE~
252 SI----HLS-D--G--RI-----YE-H----F--DHVIYCVGRSPDTENLKLEKLNVE-T 289
1ONFA
SI----HLS-D--G--RI-----YE-H----F--DHVIYCVGRSPDTENLKLEKLNVE-T
EE~~~~EEC~C~~C~~EE~~~~~EE~C~~~~E~~EEEEEECCEEEECCCCCCCCCCEE~E
1GERA
TL----ELE-D--G--R------SE-T----V--DCLIWAIGREPANDNINLEAAGVK-T
EE~~~~EEC~C~~C~~E~~~~~~EE~E~~~~E~~EEEEEECCEEEECCCCCHHHHCEE~E
1BWCA
SM----VTAVP--G--RLPVMTMIP-D----V--DCLLWAIGRVPNTKDLSLNKLGIQ-T
EE~~~~EEECC~~C~~CCEEEEEEE~C~~~~E~~EEEEEECCEEEECCCCCHHHHCEE~E
1ZDLA
QVTWEDHAS-G--K--ED-----TG-T----F--DTVLWAIGRVPETRTLNLEKAGIS-T
EEEEEEECC~C~~E~~EE~~~~~EE~E~~~~E~~EEEEEECCEEECCCCCCCCCCCEE~E
1AOGA
SV----TFE-S--G--KK-----MD------F--DLVMMAIGRSPRTKDLQLQNAGVM-I
EE~~~~EEC~C~~C~~EE~~~~~EE~~~~~~E~~EEEEEECCEEEECHHHCHHHCCEE~E
1H6VA
N-------S-E--E--TI-----ED-E----F--NTVLLAVGRDSCTRTIGLETVGVK-I
C~~~~~~~C~C~~E~~EE~~~~~EC~E~~~~E~~EEEEECCCEEEECCCCCCCCCCCC~C
1FEAA
HV----VFE-S--G--AE-----AD------Y--DVVMLAIGRVPRSQTLQLEKAGVEVA
EE~~~~EEC~C~~C~~EE~~~~~EE~~~~~~E~~EEEEEECEEEEECCCCCHHHHCEEEC
3LADA
TV----KFV-DAEG--EK-----SQ-A----F--DKLIVAVGRRPVTTDLLAADSGVT-L
EE~~~~EEE~ECCE~~EE~~~~~EE~C~~~~E~~EEEEEECEEEEECCCCCCCCCCEE~E
1LPFA
TV----TFT-DANG--EQ-----KE-T----F--DKLIVAVGRRPVTTDLLAADSGVT-L
EE~~~~EEE~CCCC~~EE~~~~~EE~E~~~~E~~EEEEEEEEEEEECCCCCCCCCCEE~E
1EBDA
VT----YEA-N--G--ET-----KTID----A--DYVLVTVGRRPNTDELGLEQIGIKMT
EE~~~~EEE~C~~C~~EE~~~~~EEEE~~~~E~~EEEEEECCEEECCCCCCHHHHCCCCC
1JEHA
EI----VVE-D--T--KT-----NK-QENLEA--EVLLVAVGRRPYIAGLGAEKIGLE-V
EE~~~~EEE~E~~E~~CC~~~~~CE~EEEEEE~~EEEEEECCEEEECCCCCCHHHCEE~E
1DXLA
TV----EPS-A--GGEQT-----II-E----A--DVVLVSAGRTPFTSGLNLDKIGVE-T
EE~~~~EEE~C~~CCCCE~~~~~EE~E~~~~E~~EEEEECCEEEEECCCCCCCCCCEE~E
1ZMCA
SI----EAA-S--G--GK-----AE-V----ITCDVLLVCIGRRPFTKNLGLEELGIE-L
EE~~~~EEE~C~~C~~CC~~~~~CE~E~~~~EEEEEEEEECCEEEECCCCCHHHHCCC~C
2A8XA
VT----VTK-D--G--VA-----QELK----A--EKVLQAIGFAPNVEGYGLDKAGVA-L
EE~~~~EEE~C~~C~~CE~~~~~EEEE~~~~E~~EEEEEECEEEEECCCCCHHHHCCC~C
1ZK7A
VL----TTT-H--G--EL--------R----A--DKLLVATGRTPNTRSLALDAAGVT-V
EE~~~~EEC~C~~C~~EE~~~~~~~~E~~~~E~~EEEEEEEEEEEECCCCCHHHHCEE~E
1XDIA
LV----TMT-D--G--RT-----VE-G-------SHALMTIGSVPNTSGLGLERVGIQ-L
EE~~~~EEC~C~~C~~CE~~~~~EE~E~~~~~~~EEEEEECEEEEECCCCCHHHCCCC~E
290 N--N-N-YIVVDENQRTSVNNIYAVGDCC---MVKKSKEIEDLNLLKLYNEERYLNKKEN 342
1ONFA
N--N-N-YIVVDENQRTSVNNIYAVGDCC---MVK-------------------------
C~~C~C~EEEEECCCCCCCCCEEEECCEE~~~EEE~~~~~~~~~~~~~~~~~~~~~~~~~
1GERA
NE-K-G-YIVVDKYQNTNIEGIYAVGDNT---GA--------------------------
CC~C~C~EEECCCCCCCCCCCEEEECCCC~~~CC~~~~~~~~~~~~~~~~~~~~~~~~~~
1BWCA
D--D-KGHIIVDEFQNTNVKGIYAVGDVC---G---------------------------
C~~C~CCEEECCCCCCCCCCCEEEECCCC~~~C~~~~~~~~~~~~~~~~~~~~~~~~~~~
1ZDLA
NPKN-Q-KIIVDAQEATSVPHIYAIGD---------------------------------
CCCC~C~EEECCCCCCCCCCCEEEECC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1AOGA
K--N-G-GVQVDEYSRTNVSNIYAIG----------------------------------
C~~C~C~EEECCCCCCCCCCCEEEEC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1H6VA
NEKT-G-KIPVTDEEQTNVPYIYAIGDIL---EGK-------------------------
CCCC~C~EEECCCCCCCCCCCEEEECCEE~~~ECC~~~~~~~~~~~~~~~~~~~~~~~~~
1FEAA
K--N-G-AIKVDAYSKTNVDNIYAIGD---------------------------------
C~~C~C~EEECCCCCCCCCCEEEEECC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
3LADA
DE-R-G-FIYVDDYCATSVPGVYAIG----------------------------------
CC~C~C~EEECCCCCCCCCCCEEEEC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1LPFA
DE-R-G-FIYVDDHCKTSVPGVFAIG----------------------------------
CC~C~C~EEECCCCCCCCCCCEEEEC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1EBDA
N--R-G-LIEVDQQCRTSVPNIFAIG----------------------------------
C~~C~C~EEECCCCCCCCCCCEEEEC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1JEHA
D--K-RGRLVIDDQFNSKFPHIKVVGD---------------------------------
C~~C~CCEEECCCCCCCCCCCEEEECC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1DXLA
D--KLG-RILVNERFSTNVSGVYAIG----------------------------------
C~~CCC~EEECCCCCCCCCCCEEEEC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1ZMCA
DP-R-G-RIPVNTRFQTKIPNIYAIGDVVAGPMLAHKAEDEGIICVE-------------
CC~C~C~EEECCCCCCCCCCCEEEECCCCCCCCCHHHHHHHHHHHHH~~~~~~~~~~~~~
2A8XA
T--D-R-KAIGVDDYRTNVGHIYAIG---------------DVNGL--------------
C~~C~C~CEEECCCCCCCCCCEEEEC~~~~~~~~~~~~~~~CCCCC~~~~~~~~~~~~~~
1ZK7A
NA-Q-G-AIVIDQGMRTSNPNIYAAGDCT-------------------------------
CC~C~C~EEECCCCCCCCCCCEEEECCCC~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
1XDIA
G--RGN-YLTVDRVSRTLATGIYAAGDCT---GL--------------------------
E~~ECC~EEECCCCCCCCCCCEEEECCCC~~~CC~~~~~~~~~~~~~~~~~~~~~~~~~~
343 VTEDIFYNVQLTPVAINAGRLLADRLF-LKK-TRKTNYKLIPTVIFSHPPIGTIGLSEEA 400
1ONFA
-----FYNVQLTPVAINAGRLLADRLF-LKK-TRKTNYKLIPTVIFSHPPIGTIGLSEEA
~~~~~CCCCCCHHHHHHHHHHHHHHHC~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1GERA
--------VELTPVAVAAGRRLSERLF-NNKPDEHLDYSNIPTVVFSHPPIGTVGLTEPQ
~~~~~~~~CCCHHHHHHHHHHHHHHHC~CCCCCCCCCCCCCEEEECCCEEEEEECCCHHH
1BWCA
-------KALLTPVAIAAGRKLAHRLFEYKE-DSKLDYNNIPTVVFSHPPIGTVGLTEDE
~~~~~~~CCCCHHHHHHHHHHHHHHHHHCCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1ZDLA
VAEG---RPELTPTAIKAGKLLAQRLF-GKS-STLMDYSNVPTTVFTPLEYGCVGLSEEE
EEEC~~~CCCCHHHHHHHHHHHHHHHC~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1AOGA
---DVTNRVMLTPVAINEAAALVDTVF-GTT-PRKTDHTRVASAVFSIPPIGTCGLIEEV
~~~CCCCCCCCHHHHHHHHHHHHHHHH~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1H6VA
--------LELTPVAIQAGRLLAQRLY-GGS-TVKCDYDNVPTTVFTPLEYGCCGLSEEK
~~~~~~~~CCCHHHHHHHHHHHHHHHH~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1FEAA
VTD----RVMLTPVAINEGAAFVDTVF-ANK-PRATDHTKVACAVFSIPPMGVCGYVEED
CCC~~~~CCCCHHHHHHHHHHHHHHHC~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
3LADA
---DVVRGAMLAHKASEEGVVVAERIA-GHK-A-QMNYDLIPAVIYTHPEIAGVGKTEQA
~~~CCCCCCCCHHHHHHHHHHHHHHHH~CCC~C~CCCCCCCEEEECCCEEEEEECCCHHH
1LPFA
---DVVRGAMLAHKASEEGVMVAERIA-GHK-A-QMNYDLIPSVIYTHPEIAWVGKTE--
~~~CCCCCCCCHHHHHHHHHHHHHHHH~CCC~C~CCCCCCCEEEECCCEEEEEECCCH~~
1EBDA
---DIVPGPALAHKASYEGKVAAEAI--AGH-PSAVDYVAIPAVVFSDPECASVGYFEQQ
~~~CCCCCCCHHHHHHHHHHHHHHHH~~HCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1JEHA
----VTFGPMLAHKAEEEG-IAAVEML-KTG-HGHVNYNNIPSVMYSHPEVAWVGKTEEQ
~~~~CCCCCCCHHHHHHHH~HHHHHHH~HCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1DXLA
---DVIPGPMLAHKAEEDGVACVEYL--AGK-VGHVDYDKVPGVVYTNPEVASVGKTEEQ
~~~CCCCCCCCHHHHHHHHHHHHHHH~~HCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1ZMCA
--------------GMAGGAVHID-------------YNCVPSVIYTHPEVAWVGKSEEQ
~~~~~~~~~~~~~~HHHCCCCCCC~~~~~~~~~~~~~CCCCEEEECCCEEEEEECCCHHH
2A8XA
--------LQLAHVAEAQGVVAAETIA-GAE-TLTLGDHRLPRATFCQPNVASFGLTEQQ
~~~~~~~~CCCHHHHHHHHHHHHHHHH~CCC~CCCCCCCCCEEEECCCEEEEEECCCHHH
1ZK7A
------DQPQFVYVAAAAGTRAAINM--TGG-DAALDLTAMPAVVFTDPQVATVGYSEAE
~~~~~~CCCCCHHHHHHHHHHHHHHH~~HCC~CCEEECCCCEEEECCCEEEEEECCCHHH
1XDIA
--------LPLASVAAMQGRIAMYHAL-GEG-VSPIRLRTVAATVFTRPEIAAVGV-PQS
~~~~~~~~CCCHHHHHHHHHHHHHHHH~CCC~CCEEECCCCEEEECCCEEEEEECC~CHH
401 AIQIYGKENVKIYESKFTNLFFSVYDIEPELKEKT-YL-KLVCVGK-D-ELIKGLHIIGL 456
1ONFA
AIQIYGKENVKIYESKFTNLFFSVYDIEPELKEKT-YL-KLVCVGK-D-ELIKGLHIIGL
HHHHHCCCEEEEEEEEEECCCCCCCCCCCCCCCEE~EE~EEEEECC~C~CCCEEEEEEEC
1GERA
AREQYGDDQVKVYKSSFTAMYTAV----TTHRQPC-RM-KLVCVGS-E-EKIVGIHGIGF
HHHHCCCCCEEEEEEEEEEHHHHC~~~~CCCEEEE~EE~EEEEECC~C~CEEEEEEEECC
1BWCA
AIHKYGIENVKTYSTSFTPMYHAV----TKRKTKC-VM-KMVCANK-E-EKVVGIHMQGL
HHHHHCCCEEEEEEEEEECCCCCC~~~~CCCCCEE~EE~EEEEECC~C~CEEEEEEEEEC
1ZDLA
AVALHGQEHVEVYHAYYKPLEFTVADRD---ASQC-YI-KMVCMREPP-QLVLGLHFLGP
HHHHHCCCEEEEEEEEECCHHHHCCCCC~~~CCCE~EE~EEEEEECCC~CEEEEEEEEEC
1AOGA
ASKRY--EVVAVYLSSFTPLMHKV----SGSKYKT-FVAKIITNHS-D-GTVLGVHLLGD
HCCCC~~EEEEEEEEEEECHHHHH~~~~HCCCCCE~EEEEEEEEEC~C~CEEEEEEEECC
1H6VA
AVEKFGEENIEVYHSFFWPLEWTVPSRD---NNKC-YA-KVICNLK-DNERVVGFHVLGP
HHHHHCCCCEEEEEEEECCHHHHCCCCC~~~CCEE~EE~EEEEECC~CCCEEEEEEEEEC
1FEAA
AAKKY--DQVAVYESSFTPL---MHNISGSTYKKF-MV-RIVTNHA-D-GEVLGVHMLGD
HHHCC~~EEEEEEEEEEECC~~~HHHHHCCCCCEE~EE~EEEEEEC~C~CEEEEEEEECC
3LADA
LKAEGVAINVGVFPFAASGRAMAAND------TAG-FV-KVIADAK-T-DRVLGVHVIGP
HHHHCEEEEEEEECCCCHHHHHHHCC~~~~~~CCC~EE~EEEEEEC~C~CEEEEEEEECC
1LPFA
--QTLKAEGVEVNVGTFP---FAASGRAMAANDTT-GLVKVIADAK-T-DRVLGVHVIGP
~~HHHHHHCEEEEEEEEC~~~CCCHHHHHHHCCCC~CEEEEEEEEC~C~EEEEEEEEECC
1EBDA
A----KDEGIDVIAAKFP---FAANGRALALNDTDGFL-KLVVRKE-D-GVIIGAQIIGP
H~~~~HHHCEEEEEEEEC~~~CCCCHHHHHHCCCCCEE~EEEEEEC~C~CCCEEEEEECC
1JEHA
LKEA----GIDYKIGKFP--FAANSRAKTNQDTEG-FV-KILIDSK-T-ERILGAHIIGP
HHHH~~~~CEEEEEEEEC~~CCCHHHHHHHCCCCC~EE~EEEEEEC~C~CEEEEEEEEEE
1DXLA
V----KETGVEYRVGKFP--FMANSRAKAIDNAEG-LV-KIIAEKE-T-DKILGVHIMAP
H~~~~HHHCEEEEEEEEC~~CCCHHHHHHHCCCCC~EE~EEEEEEC~C~CEEEEEEEEEE
1ZMCA
----LKEEGIEYKVGKFP--FAANSRAKTNADTDG-MV-KILGQKS-T-DRVLGAHILGP
~~~~HHHHCEEEEEEEEC~~CCCCHHHHHHCCCCC~EE~EEEEEEC~C~CEEEEEEEECC
2A8XA
A----RNEGYDVVVAKFP--FTANAKAHGVGDPSG-FV-KLVADAK-H-GELLGGHLVGH
H~~~~HHHCEEEEEEEEC~~CCCCHHHHHHCCCCC~EE~EEEEEEC~C~CCEEEEEEEEE
1ZK7A
A------HHDGIETDSRTLTLDNVPRALANFDTRG-FI-KLVIEEG-S-HRLIGVQAVAP
H~~~~~~HHHCEEEEEEEECCCCHHHHHHHCCCCC~EE~EEEEEEC~C~CEEEEEEEEEC
1XDIA
VIDAGSVAARTIMLPLRTNARAKMSEM-----RHG-FV-KIFCRRS-T-GVVIGGVVVAP
HHHHCCEEEEEEEEECCCCHHHHHHCC~~~~~CEE~EE~EEEEEEC~C~CEEEEEEEEEC
457 NADEIVQGFAVALKMNATKKDF-DETIPIHPTAAEEFLTLQPWMK 500
1ONFA
NADEIVQGFAVALKMNATKKDF-DETIPIHPTAAEEFLTLQ~~~~
CHHHHHCHHHHHHHHCCCHHHH~HHCCCCCCCCHHHCCCCC~~~~
1GERA
GMDEMLQGFAVALKMGATKKDF-DNTVAIHPTAAEEFVTMR~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCHHHCCCC~~~~
1BWCA
GCDEMLQGFAVAVKMGATKADF-DNTVAIHPTSSEELVTLR~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCCCCCCCC~~~~
1ZDLA
NAGEVTQGFALGIKCGASYAQV-MQTVGIHPTCSEEVVKL~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCHHHHHC~~~~~
1AOGA
NAPEIIQGIGICLKLNAKISDF-YNTIGVHPTSAEELCSMR~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCCHHHCCC~~~~
1H6VA
NAGEVTQGFAAALKCGLTKQQL-DSTIGIHPVCAEIFTTL~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCHHHHHHCC~~~~~
1FEAA
SSPEIIQSVAICLKMGAKISDF-YNTIGVHPTSAEELCSMR~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCCCCCCCC~~~~
3LADA
SAAELVQQGAIAMEFGTSAEDL-GMMVFAHPALSE~~~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCH~~~~~~~~~~
1LPFA
SAAELVQQGAIGMEFGTSAEDL-GMMVFSHPTLSE~~~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCH~~~~~~~~~~
1EBDA
NASDMIAELGLAIEAGMTAEDI-ALTIHAHPTLGE~~~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCCCH~~~~~~~~~~
1JEHA
NAGEMIAEAGLALEYGASAEDV-ARVCHAHPTLSEAF~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCHHHHH~~~~~~~~
1DXLA
NAGELIHEAAIALQYDASSEDI-ARVCHAHPTMSE~~~~~~~~~~
CHHHHHHHHHHHHHCCCCHHHH~HHCCCCCCCCHH~~~~~~~~~~
1ZMCA
GAGEMVNEAALALEYGASCEDI-ARVCHAHPTLSEAF~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHH~HHCCCCCCCHHHHH~~~~~~~~
2A8XA
DVAELLPELTLAQRWDLTASEL-ARNVHTHPTS~~~~~~~~~~~~
CCCCCHHHHHHHHHCCCHHHHH~HHCCCCCCCC~~~~~~~~~~~~
1ZK7A
EAGELIQTAALAIRNRMTVQELADQLFP~~~~~~~~~~~~~~~~~
CHHHHHHHHHHHHHHCCCHHHHHHCCCC~~~~~~~~~~~~~~~~~
1XDIA
IASELILPIAVAVQNRITVNEL-AQTLAVYPS~~~~~~~~~~~~~
CHHHHHHHHHHHHHCCCCHHHH~HHCCCCCCC~~~~~~~~~~~~~
Blast e-value: 2.77315E-111
1 >gi|33861124|ref|NP_892685.1| probable glutathione reductase (NADPH) [Prochlorococcus marinus subsp. pastoris str. CCMP1378] >gnl|BL_ORD_ID|766800 probable glutathione reductase (NADPH) [Prochlorococcus marinus subsp. pastoris str. CCMP1986]
match: ----FDLI-VLGAGSGGLAAAKRAASY------GAKVAIIEV--NK---------IGGTCVI-RGCVPKKLMVYA-ANNRRN-M---LSSE--GYGL-ISK---EITFESNILLKN-VREEVSRLSVL-HSNS--LKK--LNVKVFEG-LGR-F-LNQN-------TVEVVCPK-TKNIL------RKVS--------AKS-I-LISVGGKP---K---KLNIPG---T--DFA----W--TSD--DIFE-L-K-D-F-PK-K-L-----LIVGGGYIACEFA-SI-FKN---LGTEVTQLIRGE-NL--L-NGFDKD-L--S-ECLE-KSMTSLGINL--KF--KNQLKSI-KKI-ND-G----LE---S-TLESG-SKL-------LTDNILVATGREPSLKRLN--LDTLNLKM----DGIYLE-VNELN--KT--S---ISNIFAI--GD---I---------------------------V-K----RPNLTPVAIEQG-RVF---A-DNYFAALK---R--KVNY--ENI-PKAVFTIPEISTVGLSEEKANEI-YSEVNVQVFKC-NFTPMSNTF----KKNKS-KCML--KLV-VNKKNDKVL-GCHMFGEAA--SEII-QMVAVSLNTGI--TKKDFDTTMALHPTISEEFVT-M--------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 5.01624E-110
2 >gi|136619|sp|P13110|TYTR_TRYCO Trypanothione reductase (TR) (N(1),N(8)-bis(glutathionyl)spermidine reductase) >gnl|BL_ORD_ID|880574 trypanothione-disulfide reductase (EC 1.8.1.12) - Trypanosoma congolense >gnl|BL_ORD_ID|880574 trypanothione reductase
match: ----FDLV-IIGAGSGGLEAGWNAATLYK-----KRVAVVDVQTVHGPPF-FA-ALGGTCVN-VGCVPKKLMVTG-AQYMDQ-LRE-SA----GFGWEFDA--STIKANWKTLIAA-KNAAVLDINKS-YEDM--FKDT-EGLEFFLG-WGA-L-EQKN--------V-VTVRE-GAD------PKSKVKER----LQAEH-I-IIATGSWP---QM---LKIPG--I---EHC----I--SSN--EAFY-L-E-E-P-PR-R-V-----LTVGGGFISVEFA-GI-FNAYKPVGGKVTLCYRNN-PI--L-RGFDYT-L--R-QELT-KQLVANGIDI----MTNENPSKI-ELN-PD-G-SKHVT-F-----ESG-KTLD-------VDVVMMAIGRLPRTGYLQ--LQTVGVNLTDK--G-AIQ-VDEFS--RT--N---VPNIYAI--GD-------------------------------VTG----RIMLTPVAINEG-ASV---V-DTIFGSKP---R--KTDH--TRV-ASAVFSIPPIGTCGLTEEEAAKS-F--EKVAVYLS-CFTPLMHNISG--SKYK--KFVA--KII-TDHGDGTVV-GVHLLGDSS--PEII-QAVGICMKLNA--KISDFYNTIGVHPTSAEELCS-MRTP-----------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.47587E-109
3 >gi|1942669|pdb|1FEA|A Chain A, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.2 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain B, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.2 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain C, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.2 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain D, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.2 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain A, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.0 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain B, Unliganded Crithidia Fasciculata Trypanothione Reductase At 2.0 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain A, Unliganded Crithidia Fasciculata Trypanothione Reductase At 1.7 Angstrom Resolution >gnl|BL_ORD_ID|275408 Chain B, Unliganded Crithidia Fasciculata Trypanothione Reductase At 1.7 Angstrom Resolution
match: ----YDLV-VIGAGSGGLEAGWNAASLHK-----KRVAVIDLQKHHGPPH-YA-ALGGTCVN-VGCVPKKLMVTG-ANYMDT-IRE-SA----GFGWELDR--ESVRPNWKALIAA-KNKAVSGINDS-YEGM--FADT-EGLTFHQG-FGA-L-QDNH-------TVLVRESA-DPN-----------SA-VLETLDTEY-I-LLATGSWP---Q---HLGIEG--D---DLC----I--TSN--EAFY-L-D-E-A-PK-R-A-----LCVGGGYISIEFA-GI-FNAYKARGGQVDLAYRGD-MI--L-RGFDSE-L--R-KQLT-EQLRANGINVR----THENPAKV-TKN-AD-G-TRHV--V-F---ESG-AEAD-------YDVVMLAIGRVPRSQTLQ--LEKAGVEVA-K-NG-AIK-VDAYS--KT--N---VDNIYAI--GD-------------------------------VTD----RVMLTPVAINEG-AAF---V-DTVFANKP---R--ATDH--TKV-ACAVFSIPPMGVCGYVEEDAAKK-Y--DQVAVYES-SFTPLMHNISG--STYK--KFMV--RIVTNHADGE-VL-GVHMLGDSS--PEII-QSVAICLKMGA--KISDFYNTIGVHPTSAEELCS-MRTP-----------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.68732E-108
4 >gi|33601404|ref|NP_888964.1| putative glutathione reductase [Bordetella bronchiseptica RB50] >gnl|BL_ORD_ID|6970 putative glutathione reductase [Bordetella bronchiseptica]
match: ----FDLY-VIGAGSGGVRAARFAAGY------GARVAVAE---SRY--------LGGTCVN-VGCVPKKLLVYG-AHFHED-FEQ-AA----GFGWNPG----RPEFDWPTLIAN-KNREIERLNGI-YRNL--LVN--SGVALHEG-HAR-I-VDPH-------TVEI---N-GQRH----------S--------AQH-I-LVATGGWP---FVP---DIPG---K--EHA----I--TSN--DAFF-L-P-E-L-PR-R-V-----LVVGGGYIAVEFA-SI-FNG---LGAQTVQVYRRE-LF--L-RGFDGS-V--R-EHLR-DELVKKGLDL--RF--NTDVERI-DKR-AD-G-V--LA-V---TLSDG-SVLET-------DCVFYATGRRAMLDDLG--LENTGVRLA-E-SG-YIE-VDDEY--RT--S---EPSILAI--GD---V---------------------------I-G----RVPLTPVALAEG-MAV---A-RRLFRPQEY--R--KVDY--DLI-PTAVFSLPNIGTVGLTTEQALER--GF-RVTRYES-RFRPMKLTL----TQSQE-KTLM--KLV-VDADSQKVL-GCHMVGPDA--GEIV-QGLAVALKAGA--TKQVFDDTIGIHPTAAEEFVT-MRTAT----------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.2359E-107
5 >gi|100037|pir||S22384 dihydrolipoamide dehydrogenase (EC 1.8.1.4) precursor - garden pea
match: -----DVV-IIGGGPGGYVAAIKAAQL------GFKTTCIE----------KRGALGGTCLN-VGCIPSKALLHS-SHMYHE-AKHSFA-N-HGV--KVS----NVEIDLAAMMGQ-KDKAVSNLT----RGIEGLFKK-NKVTYVKG-YGK-F-VSPS-------EISVDTIE-GENTV----------------VKGKH-I-IIATGSDVK--SLPG-VTID-------EKK----I-VSST--GALA-L-S-E-I-PK-K-L-----VVIGAGYIGLEMG-SV-WGR---IGSEVTVVEFAS-EI--V-PTMDAE-I--R-KQFQ-RSLEKQGMKF--KL--KTKVVGV-DTS-GD-G----VK-L---TVEPS-AGGEQTIIE--ADVVLVSAGRTPFTSGLN--LDKIGVE-TDK-LGR-IL-VNERF--ST--N---VSGVYAI--GD---V---------------------------IPG-----PMLAHKAEEDG-VAC---V-EYL-AGK---VG--HVDY--DKV-PGVVYTNPEVASVGKTEEQ-VKE-TG---VE-YRVGKF-PFMAN-SRAKAIDNA-EGLV--KII-AEKETDKIL-GVHIMAPNA--GELIHEA-AIALQYDA--SSEDIARVCHANPTMSEAI------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 7.42985E-102
6 >gi|26333601|dbj|BAC30518.1| unnamed protein product [Mus musculus]
match: ----FDYL-VIGGGSGGLASARRAAEL------GARAAVVE---SHK--------LGGTCVN-VGCVPKKVMWNT-AVHSEF-MHD-HV-D-YGFQSCEG------KFSWHVIKQK-RDAYVSRLNTI-YQNN--LTK--SHIEIIHG-YAT-F-ADGPRP-----TVEV---N-GKK----------FT--------APH-I-LIATGGVP---TVPHESQIPG---A--SLG----I--TSD--GFFQ-L-E-D-L-PS-R-S-----VIVGAGYIAVEIA-GI-LSA---LGSKTSLMIRHD-KV--L-RNFDSL-I--S-SNCT-EELENAG--V--EVLKFTQVKEV-KKT-SS-G-L-ELQ-V-V-TSVPG-RKPTTTMIPDV-DCLLWAIGRDPNSKGLN--LNKVGIQ-TDE-KG-HIL-VDEFQ--NT--N---VKGVYAV--GD---V---------------------------C-G----KALLTPVAIAAG-RKL---A-HRLFECKQD--S--KLDY--DNI-PTVVFSHPPIGTVGLTEDEAVHK-YGKDNVKIYST-AFTPMYHAV----TTRKT-KCVM--KMVCANKE-EKVV-GIHMQGIGC--DEML-QGFAVAVKMGA--TKADFDNTVAIHPTSSEELVT-LR-------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 8.29477E-102
7 >gi|22995260|ref|ZP_00039739.1| COG0508: Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide acyltransferase (E2) component, and related enzymes [Xylella fastidiosa Dixon]
match: -------V-VLGAGPGGYTAAFRAADL------GLDTVLVE----RYP------NLGGVCLN-VGCIPSKALLHA-AAVID---EAAHAST-FGIDF---G-KPKITLD--TLREY-KQNVVNKLT----AGLAVMAKQ-RKVRTVTG-IAH-F-VSPN-------TLDITAAD-GSTQ----------------RLHFQQ-C-IIATGSHPV--KLP---NFPW--DD-PRI-----M--DST--DALE-L-A-E-V-PK-K-L-----LVVGGGIIGLEMA-TV-YNA---LGSHVTIVEFMD-QI--I-PGTDKD-L--V-KPLA-DRMKKQGIEI--HL--NTKASHV--K--AD-K-KG-IT-V-SFEA-PT-QGTQPALKTSTYDRVLVAVGRTPTGNNIS--AEKAGVNVTER--G-FIP-VDRQM--RS--N---VPHIFAI--GD-------------------------------IVG----NPMLAHKATHEG-K-L---A-AEVAAAETESGK--HREWVARVI-PSVAYTNPEIAWIGMTETEAKAK--GL-NIGV--A-KF-PWVAS-GRAIGIGRT-EGFT--KLI-FDEDTHRII-GGAIVGVHA--GDLLAE-IGLAIEMGA--EAEDISHTIHAHPTLSESI------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.29054E-96
8 >gi|15807510|ref|NP_296246.1| 2-oxo acid dehydrogenase, lipoamide dehydrogenase E3 component [Deinococcus radiodurans] >gnl|BL_ORD_ID|372655 dihydrolipoamide dehydrogenase (EC 1.8.1.4) DR2526 [similarity] - Deinococcus radiodurans (strain R1) >gnl|BL_ORD_ID|372655 2-oxo acid dehydrogenase, lipoamide dehydrogenase E3 component [Deinococcus radiodurans]
match: ----FDVL-VIGGGPAGYVAAIRAAQL------GFKTACVD----AFERG-GKPSLGGTCLN-VGCIPSKALLDS-SERFEM-ISHDTA-E-HGI--QVAQ-P---QIDVSKMLAR-KDGVVDKLTG----GVAFLFRK-NKVTSFHG-YGK-L-VRQD---GQEWVVD---AA-GTE------------------VKAKN-V-IVATGSNPR--ALPG-VPFGG-------HI----V-ENS---GALQ-I-D-Q-V-PG-K-L-----GVIGAGVIGLELG-SV-WRR---LGAQVTILEAMP-GF--L-MAADDA-I--A-KEAL-KQFQKQGLDF--HF--GVKIGEI--KQ-DDSG----VT-V-TYE-ENG-SQVTAQ-----FDKLIVSIGRVPNTGGLN--ADGVGLAL-DE-RG-FVK-VDQHY--RT--N---LPGVYAI--GD-------------------------------VIG----GAMLAHKAEEEG-VAL---A-E-MIAGQA---G--HVNY--DVI-PWIIYTSPEIAWAGLTEKQAKER--G---LNV-KTGQF-PFSAN-GRALGHGDP-RGFV--KVV-ADAQTDKLL-GVHMIGGGV--SELIGEIVAI-MEFGG--SAEDLARTIHAHPTLSEVVKE----------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.51267E-95
9 >gi|15826394|pdb|1JEH|A Chain A, Crystal Structure Of Yeast E3, Lipoamide Dehydrogenase >gnl|BL_ORD_ID|355692 Chain B, Crystal Structure Of Yeast E3, Lipoamide Dehydrogenase
match: ----HDVV-IIGGGPAGYVAAIKAAQL------GFNTACVE----------KRGKLGGTCLN-VGCIPSKALLNN-SHLFHQMHTE--AQK-RGIDVNGD-----IKINVANFQKA-KDDAVKQLTG----GIELLFKK-NKVTYYKGN-GS-F-EDETK-------IRVTPVD-GLEG----------TVKEDHILDVKN-I-IVATGSEVT--PFPG-IEID------EE----K-I-VSST--GALS-L-K-E-I-PK-R-L-----TIIGGGIIGLEMG-SV-YSR---LGSKVTVVEFQP-QIG---ASMDGE-V--A-KATQ-KFLKKQGLDF--KL--STKV--ISAKR-NDDK-NV-VE-IVVEDTKTN-KQ-ENLE----AEVLLVAVGRRPYIAGLG--AEKIGLEV-DK-RGRLV--IDDQF--NS--K---FPHIKVV--GD-------------------------------VT----FGPMLAHKAEEEG-IAA---V-EMLKTGH----G--HVNY--NNI-PSVMYSHPEVAWVGKTEEQ-LKE-AG---ID-YKIGKF-PFAAN-SRAKTNQDT-EGFV--KIL-IDSKTERIL-GAHIIGPNA--GEMIAEA-GLALEYGA--SAEDVARVCHAHPTLSEAFKE----------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.08518E-93
10 >gi|1706442|sp|P31052|DLD2_PSEPU Dihydrolipoamide dehydrogenase (E3 component of 2-oxoglutarate dehydrogenase complex) (LPD-GLC) (Glycine oxidation system L-factor) >gnl|BL_ORD_ID|887120 lipoamide dehydrogenase
match: ---KFDVV-VIGAGPGGYVAAIKAAQL------GLKTACIEKYTDA---E-GKLALGGTCLN-VGCIPSKALLDS-SW---K-YKE--AKE--SFNVHGIS-TGEVKMDVAAMVGR-KAGIVKNLTGG-VAT---LFKA-NGVTSIQG-HGK-L-LAGKK-------VEVTKAD-GTTEV----------------IEAEN-V-ILASGSRPI--DIPP-APVD------QNVI----V--DST--GALE-F-Q-A-V-PK-R-L-----GVIGAGVIGLELG-SV-WAR---LGAEVTVLEALD-TF--L-MAADTA-V--S-KEAQ-KTLTKQGLDI--KLGARVTGSKV------N-G-N-EVE-V-TYTNAEG----EQKI---TFDKLIVAVGRRPVTTDLL--AADSGVTI-DE-RG-YIF-VDDYC--AT--S---VPGVYAI--GD---V---------------------------VRG-----MMLAHKASEEG-IMV---V-ERIKGHK----A--QMNY--DLI-PSVIYTHPEIAWVGKTE-QALKA-EGVE-VNV--G-TF-PFAAS-GRAMAANDT-GGFV--KVI-ADAKTDRVL-GVHVIGPSA--AELVQQG-AIAMEFGT--SAEDLGMMVFSHPTLSEAL------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.24596E-90
11 >gi|23049840|ref|ZP_00076877.1| COG1249: Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes [Methanosarcina barkeri]
match: ----YDII-IIGTGTAGRTFAGKVAH------SGLKIAIVD---SR--------EYGGTCPI-RGCDPKEVL----TNITKV-I-DSSNRL-MGKGVRIDA-P--LKLDWTSLIKF-KKTFTEGYSS---KTEKHLVGM--GIDTYHG-RAH-F-ENQNTV-----VVGMDKLK-G-----------------------EH-I-FLATGAKPR--TL----NIPG--E---EYI----I--TSE--KLME-T-K-K-L-PE-K-I-----IFIGGGYISMEFA-HI-ARR---TGAEVTILQRSE-KV--L-RAFDSD-M--V-DLLM-KASKAAGIKI---L-TNKPVISV-EKD-NN-G-LI-LK-V-----QSQ-SETEPEIQIFRADMVVQGAGRVADIEDLR--LENARIK-TEK--G-AIV-VDKHM--RT--S---NSRIYA--GGDCA-----------------------------LEG-----MQLTPVAALQG-EVA---A-INVLEGDS---A--EADY--TGI-PSAVFTIPVLASVGISVAKDDDK-Y---KV-IFQD---RSNWYT-----TRKKG-MEFAASKVI-IDEANDRIM-GAHILGPNA--EEAI-NIFASVMRLGL--KASDIKKLIFTYPTTCSDIPY-M--------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 6.4378E-84
12 >gi|2791905|emb|CAA73495.1| ORF454 [Staphylococcus sciuri]
match: ---KFDVV-FIGSG----HAAWHAA-L-TLKHAGKSVAIIE-----------KDTIAGTCTN-YGC-NAKILLEG-P--YE-VLEE--ASH-YP---QIIE-SDQLHVNWENLMQY-KKAVINPLS----NTLKSMFEQ-QGIEVIMGA-GK-L-VDAH-------TIDV---E-GTP------------------IQADN-I-VIATGQHSN--KL----DIEG--SA-LTH--------DSR--DFLS-L---DKM-PN-N-I-----TFIGAGIISIEFA-SIAIKS----GAEVHVIHHTD-K--PL-EGFNEKHI--A-KLIH-K-LESEGVQF--HF--NENVQSVQQA-----G-N-SYN-V---TTESG-LSIDT-------EYVLDATGRKPNVQNIG--LDELGIEYSEK--G--IR-VDDHL--RT--N---IHNIYA--SGD-------------------------------VLDKTIPK--LTPTATFES-NYI---A-SHILGINP---N--PIQY--PAI-PSVLYSLPRLSQIGVTVKEA------EDN-EAYTIKDI-PFGKQM---VFEYKN-EIEAEMTII-LNAD-KQLV-GAEIYADDA--ANLI-NLLTFIVNQKL--TAKDLNQLIFAFPGSSSGVLD-L--------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.54637E-83
13 >gi|30248843|ref|NP_840913.1| merA; mercuric reductase [Nitrosomonas europaea ATCC 19718] >gnl|BL_ORD_ID|803871 merA; mercuric reductase [Nitrosomonas europaea ATCC 19718]
match: ------VA-VIG--SGG--AAM-AAAL-KAVEQGAKVTLIE-----------RGTIGGTCVN-VGCVPSKIMIRA-AHIAH--LRRESPFD-GGIPATAP------AIDRSKLLAQ-QQARVDELRHAKYEGI--LDGN-PAITVLHGE-AR-F-KDDQSL-----VVRL-NAG-GERVVVF-----------------DR-C-LVATGASP---AVP---PIPG--LK--ESP----Y-WTST--EAL--V-S-DII-PE-R-L-----AVIGSSVVALELA-QA-FAR---LGSQVTILARST-LF--F-RE-DPA-I--G-EAIT-AAFRAEGIKV---L-EHTQASQV--AH-VD-G---EF--V-LTTAR-G----EIH-----ADKLLVATGRTPNTRSLA--LEAAGVAV-NA-QG-AIV-IDKGM--RT--S---APHIYAA--GDC-------------------------------TD----QPQFVYVAAAAG-TRA---A-INMTGGD----A--TLDL--TAM-PAVVFTDPQVATVGYSEAEA-----HHDGIET-DSRTLT-LD-NVPRALANFDT-RGFI--KLV-IEEGSGRLI-GVQAVAPEA--GELI-QTAALAIRNRM--TAQELADQLFPYLTMVEGL------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.86745E-82
14 >gi|22124238|ref|NP_667661.1| putative oxidoreductase [Yersinia pestis KIM] >gnl|BL_ORD_ID|1063114 putative oxidoreductase [Yersinia pestis KIM]
match: -----------------------------LVKQGARVAVIERY----------NNVGGGCTH-WGTIPSKALRHAVSRIIE--FNQ---NPLYS-----DN-ARTIKSSFADILNH-ADRVINQ-QTRMRQG---FYDR-NHCHMFSGD-AS-F-IDAN-------TVNVRYAD-GTSD----------------TLQADN-I-VIATGSRPY--R-P--VNVD---FN-HE----R-I-YDSD--TILQ-L-S-HE--PQ-H-V-----IIYGAGVIGCEYA-SI-FRG---LSVKVDLINTRD-RL--L-AFLDQE-M--S-DALS-YHFWNNGVVIRH----NEEFEQI-EGT-TD-G----VI-VHL---KSG-KKVK-------ADCLLYANGRTGNTSGLG--LENIGLE-ADS-RG-LLK-VNSMY--QTALS-----HVYAV--GD-------------------------------VIGY----PSLASAAYDQGRIA----A-QAMIKGE----ANVHL---IEDI-PTGIYTIPEISSVGKTEQELTAM---KVPYEVGRA-QFKHL----ARAQIVGMD-TGSL--KIL-FHRETKQIL-GIHCFGERA--AEIIHIGQAIMEQKGEGNTLEYFVNTTFNYPTMAEAY------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.88755E-81
15 >gi|23055499|ref|ZP_00081599.1| COG1249: Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes [Geobacter metallireducens]
match: ----HDLV-ILGSGSTAFAAALRAQSY------GTRVLMVEKGVP-----------GGTCIN-WGCVPSKTLIHA-ALFYQE-GK-LGAR--LGLG---EC-GGTVVLE--RLMAR-KDQVVGHLRQT--KYLDILQDV-PGLQLVKGT-GR-F-LGPDR-------LEV-----GDREI-----------------RSER-F-LVAVGGDP---RVP---RIPG--LE-STPF----L--TSR--GTLL-L-K-T-I-PQ-S-L-----VIIGGGVIAVEMG-QM-FQR---LGAKVTILEHGP-RI--L-GPVEPE-P--A-LAVR-DFLRAEGMKIVCR-------TTICLAA-QD-G-AG-VR-V-----EAE-R--DGERVSFTAEKLLVATGTAPATNGIG--LELAGVE-TDP-RG-FVT-VDERM--RT--T---APGIWAA--GDC-------------------------------TG----GMMIATVGAREG-IVA---V-DDML--NPGCGC--SMDFLSA---PMAIFTDPEVGMVGHTEEGAKAA--GFD-VVV----NVMPV-AAIPKAHVTGHT-AGVI--KMV-ADKATGRLL-GVHLACHRG--ADIINEA-ALAIRFRA--TVEDLANALHVYPSMGE---G-LR-------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 9.68744E-79
16 >gi|27467283|ref|NP_763920.1| mercuric reductase-like protein [Staphylococcus epidermidis ATCC 12228] >gnl|BL_ORD_ID|538514 mercuric reductase-like protein [Staphylococcus epidermidis ATCC 12228]
match: ---NYDLI-IIGFGKAGKTLAAHAAG------HGQKVAVVE--QSTK-------MYGGTCIN-IGCIPTKTL-----------IHD-------GIEGN--------SFKES--ITR-KKEVVQALNNKNYQG---LNSK-NNIDVLN-YNAK-F-ISNE-------IIELQDSN-GTIQ-------ETIT--------ADK-I-LINTGSRA---NIP---DIKG--ID-TAQN----I-YDST--GLLN-I---DYQ-PQ-E-L-----VIIGGGYIALEFA-SM-FAN---FGTHVTILERGD-AI--M-TNEDQD-I--A-NLIV-KDLQDKGVTI------NTNTSTI--AF-SN-N-KDQ-T-I-IHT-NHG----EIS-----ADTVLLATGRKPNTNHLG--LENTDVKI-GK-QGEVI--VNKHL--QS--T---VKHIYAA--GD-------------------------------VKG----GLQFTYISLDDY-RII---K-SHLFGDGS---R--TTE-NRGAI-PYTVFIDPPLSRVGLIASEA-KL-QGYD---ILDNKVFVS---NIPRHKINNDS-RGLF--KAV-INKDTKEIL-GASLYGKES--EELI-NLIKLAIDQHI--PYTVLRDNIYTHPTMTESFND-LFLE-----------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.34354E-76
17 >gi|28870285|ref|NP_792904.1| glutathione reductase [Pseudomonas syringae pv. tomato str. DC3000] >gnl|BL_ORD_ID|618340 glutathione reductase [Pseudomonas syringae pv. tomato str. DC3000]
match: ----FDLF-VIGAGSGGVRAARFAAGF------GARVAVAE---SRY--------LGGTCVN-VGCVPKKLLVYG-AHFSED-F-D-HAK---GFGWSLG----EASFDWSTLIAN-KDREINRLNDI-YRKL--LVD--SGVTLLEG-HAK-I-V-GPQ------QVEI---N-GQTH----------S--------AER-I-LIATGGWP---QVP---DVPG---R--EHA----I--TSN--EAFY-L-K-A-L-PK-R-V-----VVVGGGYIAVEFA-SI-FNG---LGADTTLVYRGE-LF--L-RGFDGS-V--R-SHLH-EELLKRHMTI--RF--NSDIERI-DKQ-SD-G-S--L--L-L-SMKGG-GTLET-------DCVFYATGRRPMLDNLG--LDSVDVKL-DE-HG-YIK-VDEHY--QT--S---EPSILAL--GD---V---------------------------I-G----GVQLTPVALAEG-MAV---A-RRLFKPEQY--R--PVDY--NHI-PTAVFSLPNIGTVGLTEEDAIKA--GHD-VQVFES-RFRSMKLTL----TDDQE-RTLM--KLV-VDAKTDRVL-GCHMVGPDA--GEIV-QSLAIAIKAGA--TKQVFDDTIGVHPTAAEEFVT-MR-------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.25544E-71
18 >gi|6714837|gb|AAF26175.1| glutathione reductase [Glycine max]
match: ----FDRF-TIGAGSGGVRARRFAANY------GASVAICELPFSTISSE-TTGGVGGTCVI-RGCVPKKLLVYA-SKFSHE-FEE--SN---GFGWRYDS---EPKHDWSSLIAN-KNAELQRLTGI-YKNI--LNN--AGVKLIEG-HGK-I-IDPH-------TVDV---N-GK--L--------YS--------AKH-I-LVTVGGRP---FIP---DIPG---N--EYA----I--DSD--AALD-L-P-T-K-PE-K-I-----AIVGGGYIALEFA-GI-FNG---LKSEVHVFIRQK-KV--L-RGFDEE-I--R-DFVS-EQMSVRG--I--EFHTEESPQAI-TKS-AD-G-S--FS-L-K-TNKG---TVDG------FSHIMFATGRRPNTQNLG--LESVGVKIAKD--G-AIE-VDEYS--QT--S---VPSIWAV--GD---V----------------------------TN----RINLTPVALMEG-GAL---V-KTLFQDNPT--K--P-DY--RAV-PSAVFSQPPIGQVGLTEEQAVQQ-YG--DIDIFTA-NFRPLKATL----SGLPD-RVFM--KLV-VCAKTNEVL-GLHMCGDDA--PEIV-QGFAVALKARL--TKADFDATVGIHPSAAEEFVT-MR-------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 5.41454E-71
19 >gi|37522440|ref|NP_925817.1| unknown protein [Gloeobacter violaceus] >gnl|BL_ORD_ID|337631 glr2871 [Gloeobacter violaceus PCC 7421]
match: ---KADVI-VIGSGQGGVPLAVDQAR------SGRRVVLFE-----------RGALGGSCIN-YGCTPSKALLAA-AHAAGR-AR--LAAP-LGIHAEV-------TVDFARVMER-VRGIRASFRQG-IEQ--RLAD--AGVQIVHAE-AS-F-AGSS-------TV-V---G-GGVEV-----------------QAPL-V-VINTGTGP---TIP---ELPG--LA-GLPL----L--TNL--NIFD-L---ETL-PR-C-T-----LILGGGYIGLEL--G---QGLARLGSEVHIVHNHE-RL--L-NHEEPE-V--G-EVLA-EALRRDGIHL--HLQSEVS-QAVYQ----D-G-ICALT-L-----EDG-QILRGE------AALLAAAGRTPNTAALG--APAAGIAL-DK-QG-YVA-IDEHF--RT--T---AAGVYAI--GD-------------------------------VAR----QPAFTHVSWEDY-RRL-----QAILGGEE--RT--RSD---RVL-GYAVYTEPQVGRVGLTLDQALAK--GHRARSV----TL-PMAH-IARAIEWGHD-LGFY--RLV-VDEDTDKIL-GATLVGYEA--AEIVHVLLA-HMQADS--TWKVLEQSVHIHPTYCEALPS-LARLLK---------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.89677E-70
20 >gi|6650836|gb|AAF22039.1| glutathione reductase [Candida albicans]
match: ----FDYL-VIGGGSGGVASARRAAKY------GAKVLLIE---SNFKK------FGGTCVN-VGCVPKKVMWYT-ADLAHK-KHDLYA---YGLDKEPES-IKYGDFDWAKLKHK-RDAYVTRLNGI-YENN--LKR--EKVDYAYG-FAK-F-INS-EG-----EVEVTL-S-GDQELPFLDEGKTYKKGEKLVFSADM-T-LIATGGTA---IVPP--SVPG---A--ELG----T--TSD--GFFA-L-E-K-Q-PK-K-V-----AIVGAGYIGVELS-GV-FSS---LGSETHFFIRGD-TV--L-RSFDEV-I--Q-NTVT-DYYID---NLGINIHKQSTITKI-EGS-KD-G-K---K-V-V-HLKDG-TSVE------V-DELIWTVGRKSLID-IG--LDKVGVKINDK-Q--QIV-ADEYQ--VT--N---NPKIFSL--GD-------------------------------VVG----KVELTPVAIAA----V---S-SIDIVSD-C--K--IMDI---HI-HQCILT-SRAG-LWFITREPLKI-RG-GNLKYIIL-NLSPMYYAM-------------M---MI---KKINHLS-STRSFVSDP--EE------KV-IKMGA--TKKDFDNCVAIHPTSAEELVT-M--------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.3264E-59
21 >gi|29833890|ref|NP_828524.1| putative oxidoreductase [Streptomyces avermitilis MA-4680] >gnl|BL_ORD_ID|912722 putative oxidoreductase [Streptomyces avermitilis MA-4680]
match: --IAYDVV-VLGAGPVGENVADRTRA------AGLSTAVVE-----------SELVGGECSY-WACMPSKALLRPV-------IARADARRVPGVS----H-LAQGHLDTAAVLAH-RDYEVSHWKDD--GQVGWL-DG-IGADLYRG-HGR-LS-GPR-------EVTVTGPD-GDRRV----------------LTARHAV-AVCTGSRAL---LP---DLPGLD-Q-VEP-------WTSR--EA----TS-AQAAPG-R-L-----IVVGGGVVAVEMATA--WRA---LGSEVTVLVRGK-GL--LPR-MEP--F--AGELVA-EALKEAGADVR----TGTSVSAVTRE-----G-PT----VVVLT-DTG-DRIE-------ADEILFATGRAPRTDDLG--LDTVGLE-----PGSWLS-VDDSL--RVTGS----DWLYAV--GDV-------------------------NHRALLTHQGKYQARIAGAAIAA--RAS--GV--PLLESDPWGAHAATADH--AAV-PQVVFTDPEAAAVGLSLAEA-ER-AGH-HVRAVDV-EFS----SVAGAGLYADGYRG--RARMV-VDVNRETLL-GVTFVGPGV--GELIHSA-TIAV-AG-EVPIGRLWHAVPSYPTISEVWLR-L--------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.90855E-56
22 >gi|18309716|ref|NP_561650.1| NADH oxidase [Clostridium perfringens] >gnl|BL_ORD_ID|797086 NADH oxidase [Clostridium perfringens str. 13]
match: --MSKKIV-IVGGVAGG---ASTAARLRR-LDENNQIIMFER-------G-PHVSFSN-C-----CLP--YYLSGKVENHEDLV--LMTPEKF---------YSQYRID-ARVYT----EVVS------------INRKAKEVTVRN--------------------V-V---S-GQE----------------YTESYDK-L-VLSPGAKAI---VP---PIKG--IEDVNVFTVRNV-V--DIAK-LDSFIK-D---NNSKNIS-----VIGGGFIGVEVAENLHEA-----GYNVTLIEAMD-QI--M-KPFDYDMV----QVLH-KEMYDKGVN----LIVGDKVSSF------ESG---KV--V----LESG-KKIN-------SDAVVMAIGVAPETD-LA---REAGLEIGQ--TG-AIK-VNQNY--LTNDK-----DIYAV--GDAVEV-YNS-----------------------LTN-SISRLPLAGPAQKQA-RAV---A-DHI-HGRPSRNSGY---IGSSCVQ---IF-HYNGASTGLTEGQIKAM-NLSLNYDVVRV---------IPQDKVGLMPGSEPLHFKLI-FEVPTGRIL-GAQAIGKGN--ADKRVDIIATLIKMGG--TVEDLKDL-----------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 5.71897E-55
23 >gi|15672768|ref|NP_266942.1| NADH oxidase [Lactococcus lactis subsp. lactis] >gnl|BL_ORD_ID|17729 NADH oxidase noxC [imported] - Lactococcus lactis subsp. lactis (strain IL1403) >gnl|BL_ORD_ID|17729 NADH oxidase [Lactococcus lactis subsp. lactis]
match: --MTEKIL-IVGGVAGGMSAATRLRRL----NENAEIIVFEK---------------GPYVSFANC---------------------------GLPYYVGG---EIA-EREKLIVQSAKALKNRFN------LE-VREN-SEVIAIDSEGKK-V------------TV-V--SN-GES-----------------YVESYDKL-ILSPGAKPL---IP---QIKGLN-QATNVFSLRNI---PDVDKIMTYLKA-KA--PKSA-------TIIGAGFIGLEMAENLA-KR----GLSVTIVEKAP-HV--L-PTIDRE-M--A--AFVNEELIKNNLSV---M---TNRGAVEFKN-DE---------ILL---DNG-ESLQ-------SDLTILSVGIQPET-SLA---KSAGIKLG--LRN-AIL-VDEHY--ET--S---VKDIYAV--GDAIVV-KNQ------------------------LGQDAL-ISLASPANRQGRQV----A-D-IISGLPIKNRGS----LGTAIVR--VF-DLQVASTGLSEFQLRGL-KINHKIVHVTANNHAGYYPDVTS-----------IVLKLI-FEPESGQIF-GAQAIGKEG--VDKRIDILSTAIKAKL--TVFDL--------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.59045E-49
24 >gi|24379549|ref|NP_721504.1| NADH oxidase (H2O-forming) [Streptococcus mutans UA159] >gnl|BL_ORD_ID|9051 NADH oxidase (H2O-forming) (EC 1.6.-.-) - Streptococcus mutans >gnl|BL_ORD_ID|9051 H2O-forming NADH Oxidase [Streptococcus mutans] >gnl|BL_ORD_ID|9051 NADH oxidase (H2O-forming) [Streptococcus mutans UA159] >gnl|BL_ORD_ID|9051 H2O-forming NADH oxidase
match: -----------------------------------------------------------------------------------------------------------------IG--KQ--ISGPQGLFYADKESLEAKGAKI-YMESPVTA-IDYDAKRV-----TALV---N-GQEH-----------------VESYEKL-ILATGSTPI---LP---PIKGAAIKEGSRDFEATL-KNLQFVKL----------------------AVVGAGYIGVELAEA--FKR---LGKEVILIDVVD-T--CLAGYYDQD-L--S-EMMR-QNLEDHGIEL--AF--GETVKAI-------EG-DGKVERIV--TDKASHD----------VDMVILAVGFRPNT-ALG----NAKLK-TFR-NGAFL--VDKKQ--ET--S---IPDVYAI--GDCATV-YDNA----------------------INDTNY--IALASNALRSG---I--VA-GHNAAGHKL--ESLGVQ-GSNGI---SIFGLNMVST-GLTQEKA-KR-FGYNP-EV---TAFTDFQKA---SFIEHDNYP--VTLKIV-YDKDSRLVL-GAQMASKED--MSMGIHMFSLAIQEKV--TIERLALL-----------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 7.8686E-48
25 >gi|14520939|ref|NP_126414.1| thioredoxin reductase [Pyrococcus abyssi] >gnl|BL_ORD_ID|145595 thioredoxin reductase (trxb) PAB0500 - Pyrococcus abyssi (strain Orsay) >gnl|BL_ORD_ID|145595 trxB thioredoxin reductase [Pyrococcus abyssi]
match: ---VWDVI-IIGAGPAGYTAAIYAARF------GLDTIII------------TKDLGG---N-MA-ITD--LIENYPGFPE------------GISG-------------SELAKR-MYEHVKKY------GVDVIFD---EVVRIDPAECA-YYEGP-------CQFEVKTAN-GKEY-------------------KGKTI-IIAVGAEPRKLHVPGEKEFTGRGVS---YCA------TCD--GPL--FV-------G-KEV-----IVVGGGNTALQEA--LYLHS---IGVKVTLVHRRD-KF----RA-D--------KILQ-DRLKQAGIPTI--L--NTVVTEIR-------G-TNKVESVVLKNVKTG-ETFEKK-----VDGVFIFIGYEPKTD-F---VKHLGI--TDE-YG-YIK-VDMYM--RTK-----VPGIFAA--GD-------------------------------ITN--VFK-QIA-VAVGQGAIAA--NS-AK-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.42837E-45
26 >gi|10120656|pdb|1F6M|A Chain A, Crystal Structure Of A Complex Between Thioredoxin Reductase, Thioredoxin, And The Nadp+ Analog, Aadp+ >gnl|BL_ORD_ID|613738 Chain B, Crystal Structure Of A Complex Between Thioredoxin Reductase, Thioredoxin, And The Nadp+ Analog, Aadp+ >gnl|BL_ORD_ID|613738 Chain E, Crystal Structure Of A Complex Between Thioredoxin Reductase, Thioredoxin, And The Nadp+ Analog, Aadp+ >gnl|BL_ORD_ID|613738 Chain F, Crystal Structure Of A Complex Between Thioredoxin Reductase, Thioredoxin, And The Nadp+ Analog, Aadp+
match: ------LL-ILGSGPAGYTAAVYAAR------ANLQPVLI------------------TGME-KGGQ-----LTTTTEV-ENWPGD--PNDLTG----------------PLLMER-MHEHATKFETEII-----FDHI-NKVDLQNR---P-FRLNGD--------------N-GE-----------------YTCDA---L-IIATGASARYLGLPSEEAFKGRGVS-ASA--------TCD--GFFY---------RN-QKV-----AVIGGGNTAVEEA--LYLSN---IASEVHLIHRRD--------GFRAEKI--LIKRLM-DKV-ENG-NII--LHTNRTLEEV--TG-DQMG----VTGVRLRDTQNS-DNIESLDVAG----LFVAIGHSPNT-AI---FEG-QLEL--E-NG-YIK-VQSGI--HGNATQTSIPGVFAA--GD-------------------------------VMDH-IYRQAITS-AGTGCMAAL--DA-ERYLDGLADA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.36917E-44
27 >gi|15616133|ref|NP_244438.1| thioredoxin reductase (NADPH) [Bacillus halodurans] >gnl|BL_ORD_ID|638307 thioredoxin reductase (NADPH) trxB [imported] - Bacillus halodurans (strain C-125) >gnl|BL_ORD_ID|638307 thioredoxin reductase (NADPH) [Bacillus halodurans]
match: ---VYDVV-IAGAGPAGMTAAVYTSR------ANLSTVMVER---------------G--------VPGGQM--ANTEDVE---------NYPGFDHILG---PELS---TKMFEHAK-----KF------GAEYAYGDIKEI-IDQGDL-K-L-------------VK----A-GNKE-----------------YKARA-V-IVATGAEYKKLGVPGEKELSG-----------RGVSYCAVCDGAF--F--------KGKEL-----VVVGGGDSAVEEA--VYLTR---FASKVTIIHRRD-QL----R---------AQKILQQRAFDNDKIEFIW----DHVVKQI--NG-TD-G---KVSSVTIEHAKTGEQQ-D-----FKTDGVFIYIGMLPLNEAVK-NLN---I-LNDE--G-YIV-TNEEM--ET--S---VPGIFAA--GD-------------------------------VREKSL-RQIVT--ATGDGSLAAQ-NV-QHYIEELAEK-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.671E-39
28 >gi|18450326|ref|NP_569197.1| pli0044 [Listeria innocua] >gnl|BL_ORD_ID|983475 pli0044 [Listeria innocua]
match: ----MKYV-IVGTSHAGF----EAVQTLLKKDSDAEIVVFE---------------AGSTASFLSC---------------------------GIQ--------------SYL-----EDISKSLDELHYANEASYKE--QGVD-IRMNTSV-IAINPET-----KEITTRTTD-GVEA----------------TESYDK-L-LLSPGG------VPG--TIPNVDDQHENIFYLRG----RNWADKV----K-NRMSSAKKAV------VVGAGYIGIEAA--IAYAQ---AGIDVTVVDFVD-SI--LPTYLDSEFT--S---LLTKHMEEKGMKIK----TGEGVKEFKVNE-NN-----EVTAVV---TDKG--TYE-------ADTVIISVGVRPNTQ----WLKDT-LTL--DGRG-FVE-VNEHM--ET--S---VKDVYAA--GDATAI-PFAP-----------------------TNDKAY-IALATNARRQGVIMAR-HA-----SGDADA-KIGRVN-GTSGL---AVF-DYKFATTGIKDANANSY-QGNVK-SVYKEDLIRP---SF-----MHDEEKVLM--KL-HYDADNGRIL-GAQLMSTY----DILQAINAVSVAIEAEWTVDQLAQ------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.92181E-38
29 >gi|34849459|gb|AAP58958.1| dihydrolipoamide dehydrogensae [Spiroplasma kunkelii]
match: -------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------LIKNKVKIETSV----KIKVI--K-----G-----KTV-VYDNSDG-KEVKL-----TSDYCLVSVGQTPVTTCF----ENIGLKIWER-KN--IE-VDEQC--KT--N---LPGVYAI--GN-------------------------------VVGCT----MLAHVDSVQG--IL--VI-DSI-KGKNV-----KMNY--NRI-PSCIYSFPEVATVGITEEQAIKA-----KIA-YKAFKF--LLSANGKAIAYGET-DGFV--KILC-DPKYGEIL-GVHIVAATA--TDMIY-GITACMETEG--TIHELAKTVHPYPIL----------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.27181E-37
30 >gi|15903366|ref|NP_358916.1| NADH oxidase [Streptococcus pneumoniae R6] >gnl|BL_ORD_ID|129575 NADH oxidase (EC 1.6.-.-) [imported] - Streptococcus pneumoniae (strain R6) >gnl|BL_ORD_ID|129575 NADH oxidase [Streptococcus pneumoniae R6]
match: --------------------------------------------------------------------------------------------------------------------WIGEQIDGAEGLFYSDKEKLEAKGAKV-YMNSPVLS-IDYDAK--------V-VTAEVEGKEHK-----------------ESYEKL-IFATGSTPILPPIEGVEIVKGNREFKATLENVQFVKLYQNAEEVINKLSD------KSQHL--DRIAVVGGGYIGVELAEA--FER---LGKEVVLVDIVD-TV--LNGYYDKDF-----TQMMAKNLEDHNIRL--AL--GQTVKAI-------EG-DGKVERL-ITDKES----FD-------VDMVILAVGFRPNTA-----LADGKIELFRN--GAFL--VDKKQ--ET--S---IPGVYAV--GDCATV-YDN------------------------ARKDTSYIALASNAVRTGIVGAY-NACGHELEG-------IGVQ-GSNGI---SIYGLHMVST-GLTLEKA-KA-AGYNATET----GFNDLQKP---EFMKHDNHE--VAIKIV-FDKDSREIL-GAQMVSHDIAIS-MGIHMFSLAIQEHV--TIDKLALT-----------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.79012E-35
31 >gi|21230307|ref|NP_636224.1| alkyl hydroperoxide reductase subunit F [Xanthomonas campestris pv. campestris str. ATCC 33913] >gnl|BL_ORD_ID|600053 alkyl hydroperoxide reductase subunit F [Xanthomonas campestris pv. campestris str. ATCC 33913]
match: ----FDVL-VVGGGPAGSAAAVYAAR------KGIRTGVA------------AERFGGQVLD--------TM---------------SIENFISVP----------ETEGPKMAAAL-EQHVRQYD----------------VDIMNLQRAE-QLIPAGA----DGLVEIKLAN-GAS------------------LKSKT-V-ILSTGARWRQMNVPGEDQYRN-----------KGVAYCPHCDGPL--F--------KGKRV-----AVIGGGNSGVEAA--IDLAG---IVAHVTLVEFDD-KL----RA-DE--V--LQRKL--RSLHN--VRII----TSAQTTEVL-------GDGQKVTGLVYKDRTGG----DTQHID--LEGVFVQIGLLPNTE-F---LRGT-VELSP--RGEII--VDDRG--QTN-----VPGVFAA--GDATTV-PYKQIVIAMGEGSKAALSAF---DYLIRSSAPVSADVAQAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.01448E-34
32 >gi|17559934|ref|NP_505112.1| nfrl (60.3 kD) (5I678) [Caenorhabditis elegans] >gnl|BL_ORD_ID|666605 Hypothetical protein F20D6.11 [Caenorhabditis elegans]
match: ---------------------------------------------------------------------------------------------------------------RL--R-KDDAF--YEE---RNVKFLLKT--SV----------IAVNHKS-----REVSL--SN-GET-----------------VVYSK--L-IIATGGN-----V-RKLQVPGSDLK--NICYLRKV-EEAN---IISNL------HPG-KHV-----VCVGSSFIGMEVASALAEKA-----ASVTVI--SN-TPEPL-PVFGSD-I--G-KGIR-LKFEEKGVKF--EL--AANV--VALRG-NDQG---EVSKVIL---ENG-KELD-------VDLLVCGIGVTPATK-F---LEGSGIKL-DN-RG-FIE-VDEKF--RTNISY-----IFAM--GDVVT------------------------APLPLWDIDSINIQHFQTAQAHGQHLG--Y---TIV-GKPQPGPI--VPYFW-----TLFF----FA-FGL--KFSGCN-QGSTK----------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.33737E-34
33 >gi|11253045|pir||T43405 probable dihydrolipoamide dehydrogenase (EC 1.8.1.4) - fission yeast (Schizosaccharomyces pombe) >gnl|BL_ORD_ID|171585 dihydrolipoamide dehydrogenase [Schizosaccharomyces pombe]
match: ----YDLC-VIGGGPGGYVAAIRGAQL------GLKTICVE----------KRGTLGGTCLN-VGCIPSKALLNN-SHIYHT-VKHDTKRR--GIDVS------GVSVNLSQMMKA-KDDSVKSLT----SGIEYLFKK-NKVEYAKGT-GS-F-IDPQ-------TLSVKGID-GAAD---------------QTIKAKN-F-IIATGSEVK--PFPG-VTIDE--KK---------I-VSST--GGPY-L-Y-QRY-PK-K-M-----TVLGGGIIGLEMG-SV-WSR---LGAEVTVVEFLP-AVG---GPMDAD-I--S-KALS-RIISKQGIKFKR----STKL--VSAKV-N--G--DSVE-VEIENMKNN-KR-ETYQT----DVLLVAIGRVPYTEGLG--LDKLGISM-DK-SNRVI--MDSEY--RT--N---IPHIRVI--GDAT-----------------------------L------GPMLAHKAEDEG-IAA---V-EYIAKGQ----G--HVNY--NCI-PAVMYTHPEVAWVGITEQKAKES--G---IK-YRIGTF-GFSAN-SRAKTNMDA-DGLV--KVI-VDAETDRLL-GVHMIGPMA--GELIGEA-TLALEYGA--SAEDVARVCHAHPTLSE--------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.15216E-30
34 >gi|16131244|ref|NP_417824.1| nitrite reductase (NAD(P)H) subunit [Escherichia coli K12] >gnl|BL_ORD_ID|34518 Nitrite reductase [NAD(P)H] large subunit >gnl|BL_ORD_ID|34518 nitrite reductase [NAD(P)H] (EC 1.7.1.4) - Escherichia coli (strain K-12) >gnl|BL_ORD_ID|34518 NADH-nitrate oxidoreductase apoprotein [Escherichia coli] >gnl|BL_ORD_ID|34518 nitrite reductase (NAD(P)H) subunit [Escherichia coli K12]
match: ---------------------------------------------------------------------------------DLLDKSDAAN-FDITV-FCE-EPRIAYDRVHLSSYFSHHTAEELS--LVR--EGFYEK-HGIKVLVGERA--ITINRQE--------KVIHSSAGR------------------TVFYDK-L-IMATGSYPW---IP---PIKGS--DTQDCFVYRTI-------EDLNAI---ESCARRSKR-----GAVVGGGLLGLEAAGA--LKN---LGIETHVIEFAP-ML--MAEQLDQ--M--GGEQLR-RKIESMGVRV----HTSKNTLEIV-----QEG----VEARKTMRFADG-SELE-------VDFIVFSTGIRPR-DKLA---TQCGLDVAP--RGGIV--INDSC--QT--SD---PDIYAI--GECASW-NNR-----------------------VFGLVAPGYKMAQVAVDH---ILG-S--ENAFEGADLSAKLKLL-----GV---------DVGGIG----DA----HGRTP----GARSYVYL-----------DESKE-IYKRLI-VSEDNKTLL-GAVLVGDTSDYGNLL-QLVLNAIEL-----------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 7.52353E-30
35 >gi|37675842|ref|NP_936238.1| putative oxidoreductase [Vibrio vulnificus YJ016] >gnl|BL_ORD_ID|995597 putative oxidoreductase [Vibrio vulnificus YJ016]
match: -MMSDPII-IIGS---GF-AAYQ---------------LV-----------------------------KSVRRLDAHI---PIQIFTADD--GAEYN--------KPDLSHVFSK-RQTAAD-L---VVKSGEAFA-TEHNVQL----HAH-TQVE--RVLTQQQQV-V--AN-GRCYPY-----------------SK--L-VFATGAQAF---VP---PMRGDGLAK--------V-MTLNSLQEYQA-AE-QPL-SRAQHV-----LVIGGGLIGVEIA--LDLATS---GKQVTVVEPNA-RL--LANLL-PEFI--ALP-LE-QQLMKHGIQL--AL--NSRVESVTVQ-----G-Q-TLA-IAL---HDG-REF-------AVDAVLCAAGLKANT-AVA---REAGLSV--E-RG--IC-VDLQL--NT--SD---PHIYAL--GDCAQ----------------------------IEGRML--PYLQPIVLSANVLA---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.21666E-29
36 >gi|15425686|dbj|BAB64316.1| lipoamide dehydrogenase [Arthrobacter globiformis]
match: ----FDIL-VLGGGSGGYAAALRAVQL------GLTVGLVE-----------KGKLGGTCLH-NGCIPTKALLHS-AELADH-ARD--SAK-YGVNVTLD------SIDINAVNAY-KDGIIA---GK-YKGLQGLIKS-KGITVIEGE-GK-L-QGTDTV-----VVNGTAYK-GKN------------------------I-VLATGSYSR--TLPG-LEIGG--K-----V----I--TSD--QALT-M---DYI-PK-S-A-----IILGGGVIGVEFA-SV-WKS---FGVDVTIVEGLP-SL--V-PNEDAT-I--V-KNFE-RAFKKRGIKFSTGVF----FQGV-EQN-DD-G----VK-V---TLVDG-KTFEA-------DLLLVAVGRGPVTANLG--YEEAGITI-DR--G-FVI-TNERL--HT--G---VGNVYAV--GD---I---------------------------VPG-----VQLAHRGYQQG-IFV---A-EEIAGLKPV--V--VEDI---NI-PKVTYSEPEIATVGYTEKAAKEK-FGEDQVQ--TQ-EYN-L--AGNGKSSILGT-SGLV--KLVR-QKD-GPVV-GVHMIGARM--GEQVGEA-QLIVNWEA--YPEDVAQLLHAHPTQNE--------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 4.4563E-29
37 >gi|24373856|ref|NP_717899.1| thioredoxin reductase [Shewanella oneidensis MR-1] >gnl|BL_ORD_ID|822078 thioredoxin reductase [Shewanella oneidensis MR-1]
match: -----NLL-ILGSGPAGYTAAVYAAR------ANLKPVMI------------------TGMQ-QGGQ-----LTTTTEV-ENWPGD--AEDLTG----------------PALMER-MQKHAEKFDT------EILFDHINEVTLTER---P-F--------------RLKGDN-GE-----------------YTCDA---L-IIATGASARYLGLESEEAFKGRGVS-ACA--------TCD--GFFY---------RN-QKV-----AVIGGGNTAVEEA--LYLSN---IAAEVHLIHRRD--------TFRSEKI--LIDRLM-DKV-ANG-NIILHL--NQTMDEVVGDA---MG----VTGLKMKSTKDG-AITDLA-----VAGVFVAIGHSPNT-GI---FEG-QLEM--N-NG-YLK-VQSGL--QGNATQTSIEGVFAA--GD-------------------------------VMDQH-YRQAITS-AGTGCMAAL--DA-ERYLDAKK-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.65059E-28
38 >gi|25010186|ref|NP_734581.1| Unknown [Streptococcus agalactiae NEM316] >gnl|BL_ORD_ID|261181 Unknown [Streptococcus agalactiae NEM316]
match: ----YDVI-VLGFGKAGKTLAAKLAT------QGKSVAMVE--------E-DDKMYGGTCIN-IGCIPTKTLLVSASK----------------------------NHDFQEAMTT-RNEVTSRLRA---KNFAMLDNK-DTVDVYNAK-AR-F-ISNK-------VVELT--G-GADK---------------QELTAD-VI-IINTGAKSV--QLP----IPGL-AD-SQH-----V-YDST--AIQE-L-A-H-L-PK-R-L-----GIIGGGNIGLEFA-TL-YSE---LGSKVTVIDSQS-RI--FAR--EEEEL--S-EMAQ-DYLEEMGISF--KL--SADIKSVQNED-ED------VV-ISF---ED-----EKL----SFDAVLYATGRKPNTEGLA--LENTDIKLTE--RG-AIA-VDEYC--QT--S---VENIFAV--GD-------------------------------VNG----GPQFTYISLDDSRIVL------NYLNGD----KDYSLK-NRGAV-PTSTFTNPPLATVGLDEKTAKEK--G---YQV-KSNSL--LVSAMPRAHVNNDL-RGIF--KVV-VDTETNLIL-GARLFGAES--HELI-NIITMAMDNKIPYTY--FQKQIFTHPTMVENF------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.48262E-26
39 >gi|32037653|ref|ZP_00135925.1| COG0492: Thioredoxin reductase [Pseudomonas aeruginosa UCBPP-PA14]
match: ------LI-ILGSGPAGYTAAVYAAR------ANLKPVVI------------------TGIQ-----PGGQL-TTTTEV------DNWPGDVEGLTG-------------PALMTR-MQQHAERFDT------EIVYD---HIHT-----AE-LQQRP---------FTLKGDS-GT-----------------YTCDA---L-IIATGASAQYLGMSSEEAFMGKGVSA-----------CATCDGFFY----------RNQVV-----CVVGGGNTAVEEA--LYLAN---IAKEVHLIHRRD-KL-------RSEKI--LQDKLFDKA--ENG-NV--RLHWNTTLDEVLGDA-S--G----VTGVRLKSTIDG-STSEL-----SLAGVFIAIGHKPNTD-L---FQG-QLEMRD---G-YLR-IHGGS--EGNATQTSIEGVFAA--GD-------------------------------VADH-VYRQAITS-AGAGCMAAL--DA-EKYL-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.1763E-26
40 >gi|19746953|ref|NP_608089.1| putative NADH oxidase/alkyl hydroperoxidase reductase [Streptococcus pyogenes MGAS8232] >gnl|BL_ORD_ID|781283 putative NADH oxidase/alkyl hydroperoxidase reductase [Streptococcus pyogenes MGAS8232]
match: ---LYDVL-VIGGGPAGNSAAIYAAR------KGLKTGLL------------AETFGGQVMETVG-I-EN-MI--------------------G----------TLYTEGPKLMAE-VEAHTKSYD----------------VDIIKAQLAT--SIEKKE------NIEVTLAN-GA------------------VLQAKTAI-L-ALGAK--WRNI----NVPG----EDEFRNKGVT-YCPHCDGPL--F--------EGKDV-----AVIGGGNSGLEAA--LDLAG---LAKHVYVLE-----F--L-PELKADKV------LQ-DR-AADTANM-------TIIKNVATKD-I-VG-DDHVTGLNYTERDSG----EDKHLD--LEGVFVQIGLVPNT---A-WLKDSGVNLTD--RGEII--VDKHG--STN-----IPGIFAA--GDCTDS-AYKQIIISMGSGATAAIGAF-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.13554E-25
41 >gi|15808973|gb|AAL08575.1| thioredoxin reductase [Mycobacterium avium subsp. paratuberculosis]
match: ---VHDVI-IIGSGPAGYTAALYTAR------AQLAPVVFE-----------GTSFGG------------ALMTT-TEV----------ENYPGFRDGITG---------PELMDQMREQAL-RF------GADLRMEDVESVSL-AGPVKS-----------------VTTAE-GE------------------TVRARA-V-ILAMGAAARYLGVPGEQDLLG-----------RGVSSCATCDGFF--F--------KDQDI-----AVIGGGDSAMEEAT--FLTR---FARSVTLVHRRE-EF----R---------ASRIMLERARANDKITIV----TNKAVEAV-------EG-SETVTGLRLRDTVTG----ETSTL--AVTGVFVAIGHDPRSE-L---VRDV-LD-TD-PDG-YVL-VQGR---TTATS---IPGVFAA--GD-------------------------------LVDRT-YRQAVT--AAGSGCAAAI-DAERWL------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.32147E-20
42 >gi|6320560|ref|NP_010640.1| Thioredoxin reductase; Trr1p [Saccharomyces cerevisiae] >gnl|BL_ORD_ID|187939 Thioredoxin reductase 1 >gnl|BL_ORD_ID|187939 thioredoxin-disulfide reductase (EC 1.8.1.9) TRR1 [validated] - yeast (Saccharomyces cerevisiae) >gnl|BL_ORD_ID|187939 Similar to Thioredoxin reductase (Swiss Prot. accession numbers P09625 and Q05741), Alkyl hydroperoxide reductase F52A protein (Swiss Prot. accession number P19480), NADH Dehydrogenase (Swiss Prot. accession number P26829), and other pyridine nucleotide-disulphide oxidoreductase class-II FAD-containing flavoproteins. [Saccharomyces cerevisiae]
match: -MVHNKVT-IIGSGPAAHTAAIYLAR------AEIKPILYE----------------GMMAN--G-IAAGGQLTTTTEI----------ENFPGFPDGLTG---------SELMDRMREQS-TKF------GTEIITETVSKVDLSSKPF-K-LWTEFN-------------ED-AEP------------------VTTDA-I-ILATGASAKRMHLP------G-----EETYWQKGISACAVCDGAVPIF--------RNKPL-----AVIGGGDSACEEAQ--FLTK---YGSKVFMLVRKD-HL----R---------ASTIMQKRAEKNEKIEI---LY-NT-V-ALEAK-----G-DGKL--LNALRIKNTKKNEET---DLPVSGLFYAIGHTPATKIV------AG-QVDTDEAG-YIKTVPGSS--LT--S---VPGFFAA--GD-------------------------------VQDSK-YRQAITS-AGSGCMAAL--DA-EKYLTSLE-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.23065E-16
43 >gi|21220950|ref|NP_626729.1| putative nitrite reductase large subunit NirB [Streptomyces coelicolor A3(2)] >gnl|BL_ORD_ID|1075320 putative nitrite reductase large subunit NirB [Streptomyces coelicolor A3(2)]
match: ---------------------------------------------------------------------------------------------------------------------------------------FIAE-HGIELRVGDPAE--TIDREAR-------RVTARS-G------------------LVVEYD-VL-VLATGSYPF---VP---PVPG---KDAEGCF---V-Y-RTIEDLLA--IE-AYARTRATTG-----AVVGGGLLGLEAAGA--LNG---LGLTSHIVEFAP-RL--MPVQVDEGGG--A--ALL-RTIEEMGLSVHTGV--GTQ--EI-TTG-ED-G-A--VTGMKL---SDG-SELAT-------DLVVFSAGVRPR-DQLA---RDCGLAVGER-GG--IS-VDERC--RTVTDE----RVFAI--GECALA-AD-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 5.24769E-10
44 >gi|396309|gb|AAC43068.1| unknown dehydrogenase A
match: -------------------------------KQGARVAVIERYQ----------NVGGGCTH-WGTIPSK-LRHAVSRIIE--FNQ---NPLYSDHSRL------LRSSFADILNH-ADNVINQ-QTRMRQG---FYER-NHCEILQGN-AR-F-VDEH-------TLALDCPD-GSVE----------------TLTAEK-F-VIACGSRPY--H-PT--DV--------DFTHPR-I-YDSD--SILS-M-H-HE--PR-H-V-----LIYGAGVIGCEYA-SI-FRG---MDVKVDLINTRD-RL--L-AFLDQE-M--S-DSLS-YHFWNSGVVIRH----NEEYEKI--EG-CDDG-------V-IMHLKSG-KKLK-------ADCLLYANGRTGNTDSLA--LQNIGLE-TDS-RGQ-LK-VNSMY--QTAQ-----PHVYAV--GD-------------------------------VIGY----PSLASAAYDQGRIA----A-QALVKGE-------ATAHLIEDI-PTGIYTIPEISSVGKTEQQLTAM---KVPYEVGRA-QFKHL----ARAQIVGMN-VGTL--KIL-FHRETKEIL-GIHCFGERA--AEIIHIGQAIMEQKGGGNTIEYFVNTTFNYPTMAEAY------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 9.26028E-10
45 >gi|19703527|ref|NP_603089.1| Sarcosine oxidase alpha subunit [Fusobacterium nucleatum subsp. nucleatum ATCC 25586] >gnl|BL_ORD_ID|534040 Sarcosine oxidase alpha subunit [Fusobacterium nucleatum subsp. nucleatum ATCC 25586]
match: --IKYDLV-IVGGGPAGLAAAVEARKNRID-----NILVIE----------RAKELGGI-LQQ--CIHNGFGLH---EFKEELTGPEYAQRF------MDQ-LFELNIEY-KL-----DTMV--L------GI-----SENKI--VQA-------INS---------V-----D-G-----------------YMIIQAKS-V-ILTMGCR---ERTRGAIAIPGDRPAG--------I-FTAGAAQRYINMEG-YMV---GKRV-----VILGSGDIGLIMARRLTLE-----GAKVLAV---A-ELMPFSGGLTRNIV---------QCLDDYDIPL---YLSHTVVDII--------G-KDRVEKVIIAKVDENKKAIHGTEIEYECDTLLLSVGLIPE-NDIS---RATGIKIDPR-TNGPI--VNELM--ET--S---IVGIFA--SGNVVHV-HDLVDFVSIESRKAGK-----SAAKYIKGE---------VA--DGEYI---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.51548E-6
46 >gi|15925390|ref|NP_372924.1| nitrite reductase [Staphylococcus aureus subsp. aureus Mu50] >gnl|BL_ORD_ID|442432 nitrite reductase [Staphylococcus aureus subsp. aureus Mu50]
match: ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------L-IFATGSKAFV--IP----VPGSTLP-SVIGWRT-I-DDTE--QMMN-IAK-T----K-KKA-----IVIGGGLLGLECARGL-LDQ----GMEVTVLHLAE-WLMEM--QLDRK----AGNMLK-ADLEKQGMKF--EM--QANTTEIL-------G-EDDVEGVKL---ADG-REI-------PADLVVMAVGIRPYTE-VA--KES-GLDV--N-RG--IV-VNDVM--QTSDS-----NVYAV--GEC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 1.10402E-5
47 >gi|30261358|ref|NP_843735.1| pyridine nucleotide-disulfide oxidoreductase, class I [Bacillus anthracis str. Ames] >gnl|BL_ORD_ID|91597 pyridine nucleotide-disulfide oxidoreductase, class I [Bacillus anthracis str. Ames]
match: ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------L-LIATGVRPV---MP---EWEGRDLQ-GVHLLKT-I-PDAE--RILKTLET-NKVED----V-----TIIGGGAIGLEMAET--F---VELGKKVRMIERND-HIGTI---YDGD-M--A-EYIY-KEADKHHIEI---L-TNENVKAF--K-----G-NERVEAV---ETDKG-T--------YKADLVLVSVGVKPNTD-F---LEGTNIR-TNH-KG-AIE-VNAYM--QT--N---VQDVYAA--GDC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 2.95592E-4
48 >gi|20809045|ref|NP_624216.1| uncharacterized NAD(FAD)-dependent dehydrogenases [Thermoanaerobacter tengcongensis] >gnl|BL_ORD_ID|164821 uncharacterized NAD(FAD)-dependent dehydrogenases [Thermoanaerobacter tengcongensis]
match: ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------L-LIGTGSTAI---VP---PWEGRNLPG--VFTLKTM-GDAE--KLLDWLSS-HT--PT-KAV------VIGGGYIGLETAEALH-SR----GFKVTVLDLAP-Q---LIPTFDKE-I--A--EIAQDALTRHGVEV--HL--NEEV--VGLEG-DEKG----VKAVV---TKNG--KFE-------ADLVIISIGVRPVSE-LA---KEAGIELGPK-N--AIL-VNERL--ET--S---IPDIYAA--GDC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
Blast e-value: 3.24006E-4
49 >gi|23016561|ref|ZP_00056315.1| COG1251: NAD(P)H-nitrite reductase [Magnetospirillum magnetotacticum]
match: ----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------L-LLATGSKPLMPPLPG-LDLPGV-VAFRD------I---ADVEKMLDA--A-E----KQQRA-----VVIGGGLLGLEAAWG--LKRR---GMPVALVHLMP-TL--MERQLDVE----AGGLLQ-KDLTERGLHF---F-TSGQTEEIM--G-AEEG-CA--KGVKL---TDG-REI-------PADLVVVAIGIRPNVD-LA---KAAGLDI--N-RG--IE-VGDDM--AT--SD---PAIFSV--GEC-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
query: --MVYDLI-VIGGGSGGMAAARRAARH------NAKVALVE--KSR---------LGGTCVN-VGCVPKKIMFNA-ASVHDI-LEN--SRH-YGFDTKF-------SFNLPLLVER-RDKYIQRLNNI-YRQN--LSK--DKVDLYEG-TAS-F-LSENRI-----LIKGTKDN-NNKDN------GPLNEE---ILEGRN-I-LIAVGNKP---VFP---PVKG---I--ENT----I--SSD--EFFN-I-K-E---SK-K-I-----GIVGSGYIAVELI-NV-IKR---LGIDSYIFARGN-RI--L-RKFDES-V--I-NVLE-NDMKKN--NI--NIVTFADVVEI-KKV-SD-K-N--LS-I----------HLSDGRIYEHFDHVIYCVGRSPDTENLK--LEKLNVE-TNN-N--YIV-VDENQ--RT--S---VNNIYAV--GDCCMV-KKSKEIEDLNLLKLYNEERYLNKKENVTEDIFYNVQLTPVAINAG-RLL---A-DRLFLKKT---R--KTNY--KLI-PTVIFSHPPIGTIGLSEEAAIQI-YGKENVKIYES-KFTNLFFSVYDIEPELKE-KTYL--KLVCVGKD-ELIK-GLHIIGLNA--DEIV-QGFAVALKMNA--TKKDFDETIPIHPTAAEEFLT-LQPWMK---------------------------------------------
|
|
|
|
Click below to view the 3D structure of a PDB homolog related to your protein.
-
1ONFA
(e-value = 0.0) - Matches to residues 2 - 641
-
1GERA
(e-value = 5.0E-97) - Matches to residues 3 - 641
-
1BWCA
(e-value = 5.0E-95) - Matches to residues 3 - 641
-
1ZDLA
(e-value = 4.0E-71) - Matches to residues 3 - 640
-
1AOGA
(e-value = 2.0E-66) - Matches to residues 2 - 641
-
1H6VA
(e-value = 8.0E-66) - Matches to residues 3 - 640
-
1FEAA
(e-value = 5.0E-62) - Matches to residues 3 - 641
-
3LADA
(e-value = 1.0E-44) - Matches to residues 3 - 635
-
1LPFA
(e-value = 7.0E-43) - Matches to residues 3 - 635
-
1EBDA
(e-value = 8.0E-42) - Matches to residues 4 - 635
-
1JEHA
(e-value = 6.0E-37) - Matches to residues 3 - 637
-
1DXLA
(e-value = 7.0E-36) - Matches to residues 4 - 635
-
1ZMCA
(e-value = 2.0E-32) - Matches to residues 4 - 637
-
2A8XA
(e-value = 1.0E-30) - Matches to residues 3 - 633
-
1ZK7A
(e-value = 9.0E-30) - Matches to residues 7 - 628
-
1XDIA
(e-value = 6.0E-18) - Matches to residues 5 - 632
|
|
|

